使用者:Atry/特徵選擇
| 機器學習與資料探勘 |
|---|
 |
在機器學習和統計學中,特徵選擇 也被稱為變量選擇 , 屬性選擇 或變量子集選擇 。它是指:為了構建模型而選擇相關特徵(即屬性、指標)子集的過程。使用特徵選擇技術有三個原因:
要使用特徵選擇技術的關鍵假設是:訓練數據包含許多冗餘 或無關 的特徵,因而移除這些特徵並不會導致丟失信息。[2] 冗餘 或無關 特徵是兩個不同的概念。如果一個特徵本身有用,但如果這個特徵與另一個有用特徵強相關,且那個特徵也出現在數據中,那麼這個特徵可能就變得多餘。[3]
特徵選擇技術與特徵提取有所不同。特徵提取是從原有特徵的功能中創造新的特徵,而特徵選擇則只返回原有特徵中的子集。 特徵選擇技術的常常用於許多特徵但樣本(即數據點)相對較少的領域。特徵選擇應用的典型用例包括:解析書面文本和微陣列數據,這些場景下特徵成千上萬,但樣本只有幾十到幾百個。
介紹[編輯]
特徵選擇算法可以被視為搜索技術和評價指標的結合。前者提供候選的新特徵子集,後者為不同的特徵子集打分。 最簡單的算法是測試每個特徵子集,找到究竟哪個子集的錯誤率最低。這種算法需要窮舉搜索空間,難以算完所有的特徵集,只能涵蓋很少一部分特徵子集。 選擇何種評價指標很大程度上影響了算法。而且,通過選擇不同的評價指標,可以吧特徵選擇算法分為三類:包裝類、過濾類和嵌入類方法[3]
包裝類方法使用預測模型給特徵子集打分。每個新子集都被用來訓練一個模型,然後用驗證數據集來測試。通過計算驗證數據集上的錯誤次數(即模型的錯誤率)給特徵子集評分。由於包裝類方法為每個特徵子集訓練一個新模型,所以計算量很大。不過,這類方法往往能為特定類型的模型找到性能最好的特徵集。
過濾類方法採用代理指標,而不根據特徵子集的錯誤率計分。所選的指標算得快,但仍然能估算出特徵集好不好用。常用指標包括互信息、[3]逐點互信息,[4] 皮爾遜積矩相關係數、每種分類/特徵的組合的幀間/幀內類距離或顯著性測試評分。[4][5]
過濾類方法計算量一般比包裝類小,但這類方法找到的特徵子集不能為特定類型的預測模型調校。由於缺少調校,過濾類方法所選取的特徵集會比包裝類選取的特徵集更為通用,往往會導致比包裝類的預測性能更為低下。不過,由於特徵集不包含對預測模型的假設,更有利於暴露特徵之間的關係。許多過濾類方法提供特徵排名,而非顯式提供特徵子集。要從特徵列表的哪個點切掉特徵,得靠交叉驗證來決定。過濾類方法也常常用於包裝方法的預處理步驟,以便在問題太複雜時依然可以用包裝方法。
嵌入類方法包括了所有構建模型過程中庸道德特徵選擇技術。這類方法的典範是構建線性模型的LASSO方法。該方法給回歸係數加入了L1懲罰,導致其中的許多參數趨於零。任何回歸係數不為零的特徵都會被LASSO算法「選中」。LASSO的改良算法有Bolasso[6]和FeaLect[7]。Bolasso改進了樣本的初始過程。FeaLect根據回歸係數組合分析給所有特徵打分。 另外一個流行的做法是遞歸特徵消除(Recursive Feature Elimination)算法,通常用於支持向量機,通過反覆構建同一個模型移除低權重的特徵。這些方法的計算複雜度往往在過濾類和包裝類之間。
傳統的統計學中,特徵選擇的最普遍的形式是逐步回歸,這是一個包裝類技術。它屬於貪心算法,每一輪添加該輪最優的特徵或者刪除最差的特徵。主要的調控因素是決定何時停止算法。在機器學習領域,這個時間點通常通過交叉驗證找出。在統計學中,某些條件已經優化。因而會導致嵌套引發問題。此外,還有更健壯的方法,如分支和約束和分段線性網絡。
Subset selection[編輯]
Subset selection evaluates a subset of features as a group for suitability. Subset selection algorithms can be broken up into Wrappers, Filters and Embedded. Wrappers use a search algorithm to search through the space of possible features and evaluate each subset by running a model on the subset. Wrappers can be computationally expensive and have a risk of over fitting to the model. Filters are similar to Wrappers in the search approach, but instead of evaluating against a model, a simpler filter is evaluated. Embedded techniques are embedded in and specific to a model.
Many popular search approaches use greedy hill climbing, which iteratively evaluates a candidate subset of features, then modifies the subset and evaluates if the new subset is an improvement over the old. Evaluation of the subsets requires a scoring metric that grades a subset of features. Exhaustive search is generally impractical, so at some implementor (or operator) defined stopping point, the subset of features with the highest score discovered up to that point is selected as the satisfactory feature subset. The stopping criterion varies by algorithm; possible criteria include: a subset score exceeds a threshold, a program's maximum allowed run time has been surpassed, etc.
Alternative search-based techniques are based on targeted projection pursuit which finds low-dimensional projections of the data that score highly: the features that have the largest projections in the lower-dimensional space are then selected.
Search approaches include:
- Exhaustive
- Best first
- Simulated annealing
- 這些理論包括:
- Greedy forward selection
- Greedy backward elimination[8][9][10]
- Particle swarm optimization[11]
- Targeted projection pursuit
- Scatter Search[12]
- Variable Neighborhood Search[13][14]
Two popular filter metrics for classification problems are correlation and mutual information, although neither are true metrics or 'distance measures' in the mathematical sense, since they fail to obey the triangle inequality and thus do not compute any actual 'distance' – they should rather be regarded as 'scores'. These scores are computed between a candidate feature (or set of features) and the desired output category. There are, however, true metrics that are a simple function of the mutual information;[15] see here.
Other available filter metrics include:
- Class separability
- Error probability
- Inter-class distance
- Probabilistic distance
- Entropy
- Consistency-based feature selection
- Correlation-based feature selection
Optimality criteria[編輯]
The choice of optimality criteria is difficult as there are multiple objectives in a feature selection task. Many common ones incorporate a measure of accuracy, penalised by the number of features selected (e.g. the Bayesian information criterion). The oldest are Mallows's Cp statistic and Akaike information criterion (AIC). These add variables if the t -statistic is bigger than .
Other criteria are Bayesian information criterion (BIC) which uses , minimum description length (MDL) which asymptotically uses , Bonferroni / RIC which use , maximum dependency feature selection, and a variety of new criteria that are motivated by false discovery rate (FDR) which use something close to .
Structure Learning[編輯]
Filter feature selection is a specific case of a more general paradigm called Structure Learning. Feature selection finds the relevant feature set for a specific target variable whereas structure learning finds the relationships between all the variables, usually by expressing these relationships as a graph. The most common structure learning algorithms assume the data is generated by a Bayesian Network, and so the structure is a directed graphical model. The optimal solution to the filter feature selection problem is the Markov blanket of the target node, and in a Bayesian Network, there is a unique Markov Blanket for each node.[16]
Minimum-redundancy-maximum-relevance (mRMR) feature selection[編輯]
Peng et al. [17] proposed a feature selection method that can use either mutual information, correlation, or distance/similarity scores to select features. The aim is to penalise a feature's relevancy by its redundancy in the presence of the other selected features. The relevance of a feature set S for the class c is defined by the average value of all mutual information values between the individual feature fi and the class c as follows:
- .
The redundancy of all features in the set S is the average value of all mutual information values between the feature fi and the feature fj:
The mRMR criterion is a combination of two measures given above and is defined as follows:
Suppose that there are n full-set features. Let xi be the set membership indicator function for feature fi, so that xi=1 indicates presence and xi=0 indicates absence of the feature fi in the globally optimal feature set. Let and . The above may then be written as an optimization problem:
The mRMR algorithm is an approximation of the theoretically optimal maximum-dependency feature selection algorithm that maximizes the mutual information between the joint distribution of the selected features and the classification variable. As mRMR approximates the combinatorial estimation problem with a series of much smaller problems, each of which only involves two variables, it thus uses pairwise joint probabilities which are more robust. In certain situations the algorithm may underestimate the usefulness of features as it has no way to measure interactions between features which can increase relevancy. This can lead to poor performance[18] when the features are individually useless, but are useful when combined (a pathological case is found when the class is a parity function of the features). Overall the algorithm is more efficient (in terms of the amount of data required) than the theoretically optimal max-dependency selection, yet produces a feature set with little pairwise redundancy.
mRMR is an instance of a large class of filter methods which trade off between relevancy and redundancy in different ways.[18][19]
Global optimization formulations[編輯]
mRMR is a typical example of an incremental greedy strategy for feature selection: once a feature has been selected, it cannot be deselected at a later stage. While mRMR could be optimized using floating search to reduce some features, it might also be reformulated as a global quadratic programming optimization problem as follows:[20]
where is the vector of feature relevancy assuming there are n features in total, is the matrix of feature pairwise redundancy, and represents relative feature weights. QPFS is solved via quadratic programming. It is recently shown that QFPS is biased towards features with smaller entropy,[21] due to its placement of the feature self redundancy term on the diagonal of H.
Another global formulation for the mutual information based feature selection problem is based on the conditional relevancy:[21]
where and .
An advantage of SPECCMI is that it can be solved simply via finding the dominant eigenvector of Q, thus is very scalable. SPECCMI also handles second-order feature interaction.
For high-dimensional and small sample data (e.g., dimensionality > 105 and the number of samples < 103), the Hilbert-Schmidt Independence Criterion Lasso (HSIC Lasso) is useful.[22] HSIC Lasso optimization problem is given as
where is a kernel-based independence measure called the (empirical) Hilbert-Schmidt independence criterion (HSIC), denotes the trace, is the regularization parameter, and are input and output centered Gram matrices, and are Gram matrices, and are kernel functions, is the centering matrix, is the m-dimensional identity matrix (m: the number of samples), is the m-dimensional vector with all ones, and is the -norm. HSIC always takes a non-negative value, and is zero if and only if two random variables are statistically independent when a universal reproducing kernel such as the Gaussian kernel is used.
The HSIC Lasso can be written as
where is the Frobenius norm. The optimization problem is a Lasso problem, and thus it can be efficiently solved with a state-of-the-art Lasso solver such as the dual augmented Lagrangian method.
Correlation feature selection[編輯]
The Correlation Feature Selection (CFS) measure evaluates subsets of features on the basis of the following hypothesis: "Good feature subsets contain features highly correlated with the classification, yet uncorrelated to each other".[23][24] The following equation gives the merit of a feature subset S consisting of k features:
Here, is the average value of all feature-classification correlations, and is the average value of all feature-feature correlations. The CFS criterion is defined as follows:
The and variables are referred to as correlations, but are not necessarily Pearson's correlation coefficient or Spearman's ρ. Dr. Mark Hall's dissertation uses neither of these, but uses three different measures of relatedness, minimum description length (MDL), symmetrical uncertainty, and relief.
Let xi be the set membership indicator function for feature fi ; then the above can be rewritten as an optimization problem:
The combinatorial problems above are, in fact, mixed 0–1 linear programming problems that can be solved by using branch-and-bound algorithms.[25]
Regularized trees[編輯]
The features from a decision tree or a tree ensemble are shown to be redundant. A recent method called regularized tree[26] can be used for feature subset selection. Regularized trees penalize using a variable similar to the variables selected at previous tree nodes for splitting the current node. Regularized trees only need build one tree model (or one tree ensemble model) and thus are computationally efficient.
Regularized trees naturally handle numerical and categorical features, interactions and nonlinearities. They are invariant to attribute scales (units) and insensitive to outliers, and thus, require little data preprocessing such as normalization. Regularized random forest (RRF)[27] is one type of regularized trees. The guided RRF is an enhanced RRF which is guided by the importance scores from an ordinary random forest.
Overview on metaheuristics methods[編輯]
A metaheuristic is a general description of an algorithm dedicated to solve difficult (typically NP-hard problem) optimization problems for which there is no classical solving methods. Generally, a metaheuristic is a stochastics algorithm tending to reach a global optima. There are many metaheuristics, from a simple local search to a complex global search algorithm.
Main principles[編輯]
The feature selection methods are typically presented in three classes based on how they combine the selection algorithm and the model building.
Filter Method[編輯]

Filter-based feature selection has become crucial in many classification settings, especially object recognition, recently faced with feature learning strategies that originate thousands of cues.[28] Filter methods analyze intrinsic properties of data, ignoring the classifier. Most of these methods can perform two operations, ranking and subset selection: in the former, the importance of each individual feature is evaluated, usually by neglecting potential interactions among the elements of the joint set; in the latter, the final subset of features to be selected is provided. In some cases, these two operations are performed sequentially (first the ranking, then the selection); in other cases, only the selection is carried out.[28] Filter methods suppress the least interesting variables. These methods are particularly effective in computation time and robust to overfitting.[29]
However, filter methods tend to select redundant variables because they do not consider the relationships between variables. Therefore, they are mainly used as a pre-process method.
Wrapper Method[編輯]
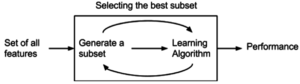
Wrapper methods evaluate subsets of variables which allows, unlike filter approaches, to detect the possible interactions between variables.[30] The two main disadvantages of these methods are :
- The increasing overfitting risk when the number of observations is insufficient.
- The significant computation time when the number of variables is large.
Embedded Method[編輯]

Recently, embedded methods have been proposed to reduce the classification of learning. They try to combine the advantages of both previous methods. The learning algorithm takes advantage of its own variable selection algorithm. So, it needs to know preliminary what a good selection is, which limits their exploitation.[31]
Application of feature selection metaheuristics[編輯]
This is a survey of the application of feature selection metaheuristics lately used in the literature. This survey was realized by J. Hammon in her thesis.[29]
| 應用領域 | 算法 | 我們的工作方式 | classifier | Evaluation Function | {0}參考號{1}:016596{/1}{/0} |
|---|---|---|---|---|---|
| SNPs | Feature Selection using Feature Similarity | 過濾 | ŕ | Phuong 2005 [30] | |
| SNPs | 這些理論包括: | {0}wrapper{/0}{1}.{/1} | Decision Tree | Classification accuracy (10-fold) | Shah 2004 [32] |
| SNPs | HillClimbing | Filter + Wrapper | Naive Bayesian | Predicted residual sum of squares | 長 |
| SNPs | Simulated Annealing | Naive bayesian | Classification accuracy (5-fold) | Ustunkar 2011 [33] | |
| Segments parole | Ants colony | {0}wrapper{/0}{1}.{/1} | Artificial Neural Network | MSE | Al-ani 2005 [來源請求] |
| 營銷 | Simulated Annealing | {0}wrapper{/0}{1}.{/1} | 回歸 | AIC, r2 | Meiri 2006 [34] |
| 經濟 | Simulated Annealing, Genetic Algorithm | {0}wrapper{/0}{1}.{/1} | 回歸 | BIC | Kapetanios 2005 [35] |
| Spectral Mass | 這些理論包括: | {0}wrapper{/0}{1}.{/1} | Multiple Linear Regression, Partial Least Squares | root-mean-square error of prediction | Broadhurst 2007 [36] |
| Microarray | Tabu Search + PSO | {0}wrapper{/0}{1}.{/1} | Support Vector Machine, K Nearest Neighbors | Euclidean Distance | Chuang 2009 [37] |
| Microarray | PSO + Genetic Algorithm | {0}wrapper{/0}{1}.{/1} | Support Vector Machine | Classification accuracy (10-fold) | Alba 2007 [38] |
| Microarray | Genetic Algorithm + Iterated Local Search | EMBEDDED | Support Vector Machine | Classification accuracy (10-fold) | Duval 2009 [31] |
| Microarray | Iterated Local Search | {0}wrapper{/0}{1}.{/1} | 回歸 | Posterior Probability | Hans 2007 [39] |
| Microarray | 這些理論包括: | {0}wrapper{/0}{1}.{/1} | K Nearest Neighbors | Classification accuracy (Leave-one-out cross-validation) | Jirapech-Umpai 2005 [40] |
| Microarray | Hybrid Genetic Algorithm | {0}wrapper{/0}{1}.{/1} | K Nearest Neighbors | Classification accuracy (Leave-one-out cross-validation) | 我 |
| Microarray | 這些理論包括: | {0}wrapper{/0}{1}.{/1} | Support Vector Machine | Sensitivity and specificity | Xuan 2011 [41] |
| Microarray | 這些理論包括: | {0}wrapper{/0}{1}.{/1} | All paired Support Vector Machine | Classification accuracy (Leave-one-out cross-validation) | Peng 2003 [42] |
| Microarray | 這些理論包括: | EMBEDDED | Support Vector Machine | Classification accuracy (10-fold) | Hernandez 2007 [43] |
| Microarray | 這些理論包括: | 混合式 | Support Vector Machine | Classification accuracy (Leave-one-out cross-validation) | Huerta 2006 [44] |
| Microarray | 這些理論包括: | Support Vector Machine | Classification accuracy (10-fold) | Muni 2006 [45] | |
| Microarray | 這些理論包括: | {0}wrapper{/0}{1}.{/1} | Support Vector Machine | EH-DIALL, CLUMP | Jourdan 2004 [46] |
| Object Recognition | Infinite Feature Selection | 過濾 | Support Vector Machine | Mean Average Precision (mAP) | Roffo 2015 [28] |
Feature selection embedded in learning algorithms[編輯]
Some learning algorithms perform feature selection as part of their overall operation. 這些心理素質包括:
- -regularization techniques, such as sparse regression, LASSO, and -SVM
- Regularized trees,[26] e.g. regularized random forest implemented in the RRF package[27]
- Decision tree[來源請求]
- Memetic algorithm
- Random multinomial logit (RMNL)
- Auto-encoding networks with a bottleneck-layer
- Submodular feature selection[47][48][49]
參見[編輯]
| 此條目已列出參考資料,但文內引註不足,部分內容的來源仍然不明。 (2010年7月) |
參考文獻[編輯]
- ^ 1.0 1.1 Gareth James; Daniela Witten; Trevor Hastie; Robert Tibshirani. An Introduction to Statistical Learning. Springer. 2013: 204.
- ^ 2.0 2.1 Bermingham, Mairead L.; Pong-Wong, Ricardo; Spiliopoulou, Athina; Hayward, Caroline; Rudan, Igor; Campbell, Harry; Wright, Alan F.; Wilson, James F.; Agakov, Felix; Navarro, Pau; Haley, Chris S. Application of high-dimensional feature selection: evaluation for genomic prediction in man. Sci. Rep. 2015, 5.
- ^ 3.0 3.1 3.2 Guyon, Isabelle; Elisseeff, André. An Introduction to Variable and Feature Selection. JMLR. 2003, 3.
- ^ 4.0 4.1 Yang, Yiming; Pedersen, Jan O. A comparative study on feature selection in text categorization. ICML. 1997.
- ^ Forman, George. An extensive empirical study of feature selection metrics for text classification. Journal of Machine Learning Research. 2003, 3: 1289–1305.
- ^ Bach, Francis R. Bolasso: model consistent lasso estimation through the bootstrap. Proceedings of the 25th international conference on Machine learning. 2008: 33–40. doi:10.1145/1390156.1390161.
- ^ Zare, Habil. Scoring relevancy of features based on combinatorial analysis of Lasso with application to lymphoma diagnosis. BMC Genomics. 2013, 14: S14. doi:10.1186/1471-2164-14-S1-S14.
- ^ Figueroa, Alejandro. Exploring effective features for recognizing the user intent behind web queries. Computers in Industry. 2015, 68: 162–169. doi:10.1016/j.compind.2015.01.005.
- ^ Figueroa, Alejandro; Guenter Neumann. Learning to Rank Effective Paraphrases from Query Logs for Community Question Answering. AAAI. 2013.
- ^ Figueroa, Alejandro; Guenter Neumann. Category-specific models for ranking effective paraphrases in community Question Answering. Expert Systems with Applications. 2014, 41: 4730–4742. doi:10.1016/j.eswa.2014.02.004.
- ^ Zhang, Y.; Wang, S.; Phillips, P. Binary PSO with Mutation Operator for Feature Selection using Decision Tree applied to Spam Detection. Knowledge-Based Systems. 2014, 64: 22–31. doi:10.1016/j.knosys.2014.03.015.
- ^ F.C. Garcia-Lopez, M. Garcia-Torres, B. Melian, J.A. Moreno-Perez, J.M. Moreno-Vega. Solving feature subset selection problem by a Parallel Scatter Search, European Journal of Operational Research , vol. 169, no. 2, pp. (2006年).
- ^ F.C. Garcia-Lopez, M. Garcia-Torres, B. Melian, J.A. Moreno-Perez, J.M. Moreno-Vega. Solving Feature Subset Selection Problem by a Hybrid Metaheuristic. In First International Workshop on Hybrid Metaheuristics , pp. 59–68, 2004.
- ^ M. Garcia-Torres, F. Gomez-Vela, B. Melian, J.M. Moreno-Vega. High-dimensional feature selection via feature grouping: A Variable Neighborhood Search approach, Information Sciences , vol. 326, pp. {0}8.{/0} {1}{/1}
- ^ Alexander Kraskov, Harald Stögbauer, Ralph G. Andrzejak, and Peter Grassberger, "Hierarchical Clustering Based on Mutual Information", (2003) ArXiv q-bio/0311039
- ^ Aliferis, Constantin. Local causal and markov blanket induction for causal discovery and feature selection for classification part I: Algorithms and empirical evaluation (PDF). Journal of Machine Learning Research. 2010, 11: 171–234.
- ^ Peng, H. C.; Long, F.; Ding, C. Feature selection based on mutual information: criteria of max-dependency, max-relevance, and min-redundancy. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2005, 27 (8): 1226–1238. PMID 16119262. doi:10.1109/TPAMI.2005.159.教學大綱
- ^ 18.0 18.1 Brown, G., Pocock, A., Zhao, M.-J., Lujan, M. (2012). "Conditional Likelihood Maximisation: A Unifying Framework for Information Theoretic Feature Selection", In the Journal of Machine Learning Research (JMLR). [1]
- ^ Nguyen, H., Franke, K., Petrovic, S. (2010). "Towards a Generic Feature-Selection Measure for Intrusion Detection", In Proc. International Conference on Pattern Recognition (ICPR), Istanbul, Turkey. [2]
- ^ Rodriguez-Lujan, I.; Huerta, R.; Elkan, C.; Santa Cruz, C. Quadratic programming feature selection (PDF). JMLR. 2010, 11: 1491–1516.
- ^ 21.0 21.1 Nguyen X. Vinh, Jeffrey Chan, Simone Romano and James Bailey, "Effective Global Approaches for Mutual Information based Feature Selection". Proceeedings of the 20th ACM SIGKDD Conference on Knowledge Discovery and Data Mining (KDD'14), August 24–27, New York City, 2014. "[3]"
- ^ M. Yamada, W. Jitkrittum, L. Sigal, E. P. Xing, M. Sugiyama, High-Dimensional Feature Selection by Feature-Wise Non-Linear Lasso. Neural Computation, vol.26, no.1, pp.185-207, 2014.
- ^ M. Hall 1999, Correlation-based Feature Selection for Machine Learning
- ^ Senliol, Baris, et al. "Fast Correlation Based Filter (FCBF) with a different search strategy." Computer and Information Sciences, 2008. ISCIS'08. 23rd International Symposium on. 978-1-4244-2108-4/08 /©2008電氣與電子工程師協會[4]
- ^ Hai Nguyen, Katrin Franke, and Slobodan Petrovic, Optimizing a class of feature selection measures, Proceedings of the NIPS 2009 Workshop on Discrete Optimization in Machine Learning: Submodularity, Sparsity & Polyhedra (DISCML), Vancouver, Canada, December 2009. [5]
- ^ 26.0 26.1 H. Deng, G. Runger, "Feature Selection via Regularized Trees", Proceedings of the 2012 International Joint Conference on Neural Networks (IJCNN), IEEE, 2012
- ^ 27.0 27.1 RRF: Regularized Random Forest, R package on CRAN
- ^ 28.0 28.1 28.2 Roffo, Giorgio; Melzi, Simone; Cristani, Marco. Infinite Feature Selection. www.cv-foundation.org. International Conference on Computer Vision. 2015 [2016-01-25].
- ^ 29.0 29.1 J. Hammon. Optimisation combinatoire pour la sélection de variables en régression en grande dimension : Application en génétique animale. 2014年11月10日
- ^ 30.0 30.1 T. M. Phuong, Z. Lin et R. B. Altman. Choosing SNPs using feature selection. Proceedings / IEEE Computational Systems Bioinformatics Conference, CSB. IEEE Computational Systems Bioinformatics Conference, pages 301-309, 2005. PMID:
- ^ 31.0 31.1 B. Duval, J.-K. Hao et J. C. Hernandez Hernandez. A memetic algorithm for gene selection and molecular classification of an cancer. In Proceedings of the 11th Annual conference on Genetic and evolutionary computation, GECCO '09, pages 201-208, New York, NY, USA, 2009. (u A/cm2)
- ^ S. C. Shah et A. Kusiak. Data mining and genetic algorithm based gene/SNP selection. Artificial intelligence in medicine, vol. 31, no. 3, pages 183-196, July 2004. PMID:
- ^ G. Ustunkar, S. Ozogur-Akyuz, G. W. Weber, C. M. Friedrich et Yesim Aydin Son. Selection of representative SNP sets for genome-wide association studies : a metaheuristic approach. Optimization Letters, November 2011.
- ^ R. Meiri et J. Zahavi. Using simulated annealing to optimize the feature selection problem in marketing applications. European Journal of Operational Research, vol. 171, no. 3, pages 842-858, Juin 2006
- ^ G. Kapetanios. Variable Selection using Non-Standard Optimisation of Information Criteria. Working Paper 533, Queen Mary, University of London, School of Economics and Finance, 2005.
- ^ D. Broadhurst, R. Goodacre, A. Jones, J. J. Rowland et D. B. Kell. Genetic algorithms as a method for variable selection in multiple linear regression and partial least squares regression, with applications to pyrolysis mass spectrometry. Analytica Chimica Acta, vol. 348, no. 1-3, pages 71-86, August 1997.
- ^ Chuang, L.-Y.; Yang, C.-H. Tabu search and binary particle swarm optimization for feature selection using microarray data. Journal of computational biology. 2009, 16 (12): 1689–1703. PMID 20047491. doi:10.1089/cmb.2007.0211.
- ^ E. Alba, J. Garia-Nieto, L. Jourdan et E.-G. Talbi. Gene Selection in Cancer Classification using PSO-SVM and GA-SVM Hybrid Algorithms. Congress on Evolutionary Computation, Singapor : Singapore (2007), 2007
- ^ C. Hans, A. Dobra et M. West. Shotgun stochastic search for 'large p' regression. Journal of the American Statistical Association, 2007.
- ^ T. Jirapech-Umpai et S. Aitken. Feature selection and classification for microarray data analysis : Evolutionary methods for identifying predictive genes. BMC bioinformatics, vol. 6, no. 1, page 148, 2005.
- ^ Xuan, P.; Guo, M. Z.; Wang, J.; Liu, X. Y.; Liu, Y. Genetic algorithm-based efficient feature selection for classification of pre-miRNAs. Genetics and Molecular Research. 2011, 10 (2): 588–603. PMID 21491369. doi:10.4238/vol10-2gmr969.
- ^ S. Peng. Molecular classification of cancer types from microarray data using the combination of genetic algorithms and support vector machines. FEBS Letters, vol. 555, no. 2, pages 358-362, December 2003.
- ^ J. C. H. Hernandez, B. Duval et J.-K. Hao. A genetic embedded approach for gene selection and classification of microarray data. In Proceedings of the 5th European conference on Evolutionary computation, machine learning and data mining in bioinformatics, EvoBIO'07, pages 90-101, Berlin, Heidelberg, 2007. SpringerVerlag.
- ^ E. B. Huerta, B. Duval et J.-K. Hao. A hybrid GA/SVM approach for gene selection and classification of microarray data. evoworkshops 2006, LNCS, vol. 3907, pages 34-44, 2006.
- ^ D. P. Muni, N. R. Pal et J. Das. Genetic programming for simultaneous feature selection and classifier design. IEEE Transactions on Systems, Man, and Cybernetics, Part B : Cybernetics, vol. 36, no. 1, pages 106-117, February 2006.
- ^ L. Jourdan, C. Dhaenens et E.-G. Talbi. Linkage disequilibrium study with a parallel adaptive GA. International Journal of Foundations of Computer Science, 2004.
- ^ Das el al, Submodular meets Spectral: Greedy Algorithms for Subset Selection, Sparse Approximation and Dictionary Selection
- ^ Liu et al, Submodular feature selection for high-dimensional acoustic score spaces
- ^ Zheng et al, Submodular Attribute Selection for Action Recognition in Video
擴展閱讀[編輯]
- Feature Selection for Classification: A Review (Survey,2014)
- Feature Selection for Clustering: A Review (Survey,2013)
- Tutorial Outlining Feature Selection Algorithms, Arizona State University
- JMLR Special Issue on Variable and Feature Selection
- Feature Selection for Knowledge Discovery and Data Mining (Book)
- An Introduction to Variable and Feature Selection (Survey)
- Toward integrating feature selection algorithms for classification and clustering (Survey)
- feature subset selection and subset size optimization.pdf Efficient Feature Subset Selection and Subset Size Optimization (Survey, 2010)
- Searching for Interacting Features
- Feature Subset Selection Bias for Classification Learning
- Y. Sun, S. Todorovic, S. Goodison, Local Learning Based Feature Selection for High-dimensional Data Analysis, IEEE Transactions on Pattern Analysis and Machine Intelligence , vol. 32, no. 9, pp. {0}8.{/0} {1}{/1}
外部連結[編輯]
- A comprehensive package for Mutual Information based feature selection in Matlab
- Infinite Feature Selection (Source Code) in Matlab
- Feature Selection Package, Arizona State University (Matlab Code)
- NIPS challenge 2003 (see also NIPS)
- Naive Bayes implementation with feature selection in Visual Basic (includes executable and source code)
- Minimum-redundancy-maximum-relevance (mRMR) feature selection program
- FEAST (Open source Feature Selection algorithms in C and MATLAB)
[[Category:{0}3{/0}{1} {/1}{0}模型選擇{/0}]]







![{\displaystyle \mathrm {mRMR} =\max _{S}\left[{\frac {1}{|S|}}\sum _{f_{i}\in S}I(f_{i};c)-{\frac {1}{|S|^{2}}}\sum _{f_{i},f_{j}\in S}I(f_{i};f_{j})\right].}](https://wikimedia.org/api/rest_v1/media/math/render/svg/3eec7b98cd9e6fc9b3b61c0ac4712a16379c8859)


![{\displaystyle \mathrm {mRMR} =\max _{x\in \{0,1\}^{n}}\left[{\frac {\sum _{i=1}^{n}c_{i}x_{i}}{\sum _{i=1}^{n}x_{i}}}-{\frac {\sum _{i,j=1}^{n}a_{ij}x_{i}x_{j}}{(\sum _{i=1}^{n}x_{i})^{2}}}\right].}](https://wikimedia.org/api/rest_v1/media/math/render/svg/0baef01e8c550ba917099a82e0ac43e826f59d37)

![{\displaystyle F_{n\times 1}=[I(f_{1};c),\ldots ,I(f_{n};c)]^{T}}](https://wikimedia.org/api/rest_v1/media/math/render/svg/e655a9d669fdf3ca7c6572563f8b5d1c1d7af44e)
![{\displaystyle H_{n\times n}=[I(f_{i};f_{j})]_{i,j=1\ldots n}}](https://wikimedia.org/api/rest_v1/media/math/render/svg/e5d1966a8fa8bd4894b5d768dcfc4da9b1caa9de)

























![{\displaystyle \mathrm {CFS} =\max _{S_{k}}\left[{\frac {r_{cf_{1}}+r_{cf_{2}}+\cdots +r_{cf_{k}}}{\sqrt {k+2(r_{f_{1}f_{2}}+\cdots +r_{f_{i}f_{j}}+\cdots +r_{f_{k}f_{1}})}}}\right].}](https://wikimedia.org/api/rest_v1/media/math/render/svg/9dfb56cea66ca7f2e039615b8c586ed355caf9e9)


![{\displaystyle \mathrm {CFS} =\max _{x\in \{0,1\}^{n}}\left[{\frac {(\sum _{i=1}^{n}a_{i}x_{i})^{2}}{\sum _{i=1}^{n}x_{i}+\sum _{i\neq j}2b_{ij}x_{i}x_{j}}}\right].}](https://wikimedia.org/api/rest_v1/media/math/render/svg/9491bc46548bd4416952e59704e78388e8726480)
