光裂合酶
| DNA光裂合酶 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
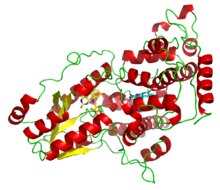 A deazaflavin photolyase from Anacystis nidulans, illustrating the two light-harvesting cofactors: FADH− (yellow) and 8-HDF (cyan). | |||||||||||
| 鑑定 | |||||||||||
| 標誌 | FAD_binding_7 | ||||||||||
| Pfam | PF03441(舊版) | ||||||||||
| InterPro | IPR005101 | ||||||||||
| PROSITE | PDOC00331 | ||||||||||
| SCOP | 1qnf / SUPFAM | ||||||||||
| |||||||||||
光裂合酶(Photolyases,EC 4.1.99.3)為一種DNA修復酵素,專門修復遭受紫外線破壞的DNA。此酵素的作用需要可見光協助活化[1]及DNA修復[2]。光裂合酶修復DNA的過程稱為光學活作用(photoreactivation)。
由系統發生學來看,光裂合酶是一個相當古老的酵素,從細菌、真菌[3],到動物體內都有[4],且在植物體內尤為重要。人類等胎盤哺乳類體內則並無此酵素活性,而是使用另一種較無效率的核苷酸切除修復法來取代光裂合酶[5]。
DNA中相鄰的胸腺嘧啶受到紫外線的照射時,會形成共價鍵,形成環丁烷橋,稱為胸腺嘧啶二聚體。形成胸腺嘧啶二聚體的基因會導致基因在複製和轉錄時出現錯誤[6]。光裂合酶會接在DNA上,切除胸腺嘧啶二聚體,並修復回原本的樣子。

光裂合酶為一種黃素蛋白,內含兩個捕光性輔因子。所有的光裂合酶都含有FADH−,並可根據他的第二輔因子分為兩類。第一類為葉酸性光裂合酶,其第二輔因子的成分為次甲基四氫葉酸蝶呤(Methenyltetrahydrofolate,MTHF),稱。第二類則為脫氮黃素性光裂合酶,其第二輔因子為8-羥-7,8-二甲基-5-脫氮核黃素(8-hydroxy-7,8-didemethyl-5-deazariboflavin,8-HDF)。FAD本身即有酵素活性,第二輔因子則可在低光狀態下加速反應進行。被光活化的光裂合酶會將FADH−的電子,轉移給胸腺嘧啶二聚體來裂解二聚體[7]。
除了第二輔因子之外,光裂合酶也可根據序列相似性分為兩種,第一種主要在革蘭氏陰性菌、陽性菌、嗜鹽古菌、真菌,和植物體內。Proteins containing this domain also include Arabidopsis thaliana cryptochromes 1 and 2, which are blue light photoreceptors that mediate blue light-induced gene expression and modulation of circadian rhythms.
、可以將光裂合酶分為兩類。The first class contains enzymes from Gram-negative and Gram-positive bacteria, the halophilic archaebacteria Halobacterium halobium,以及真菌與植物。阿拉伯芥的Cryptochrome 1與2亦含有此種結構域,作為促進藍光基因表現與調整晝夜節律的光受體。
The second class are named cryptochrome (Cry), found in species as diverse as Drosophila, Arabidopsis, Synechocystis, and Human (Cry-DASH). These were previously assumed to have no DNA repair activity because of negligible activity on double-stranded DNA. A study[4] by A. Sancar and P. Selby provided evidence to suggest this branch of cryptochromes have photolyase activity with a high degree of specificity for cyclobutane pyrimidine dimers in single-stranded DNA. Their study showed that VcCry1 from Vibrio cholerae, X1Cry from Xenopus laevis, and AtCry3 from Arabidopsis thaliana all had photolyase activity on UV irradiated ssDNA in vitro.
人類體內含光裂合酶之蛋白
[編輯]參考文獻
[編輯]- ^ Yamamoto J, Shimizu K, Kanda T, Hosokawa Y, Iwai S, Plaza P, Müller P. Loss of Fourth Electron-Transferring Tryptophan in Animal (6-4) Photolyase Impairs DNA Repair Activity in Bacterial Cells. Biochemistry. October 2017, 56 (40): 5356–5364. PMID 28880077. doi:10.1021/acs.biochem.7b00366.
- ^ Thiagarajan V, Byrdin M, Eker AP, Müller P, Brettel K. Kinetics of cyclobutane thymine dimer splitting by DNA photolyase directly monitored in the UV. Proceedings of the National Academy of Sciences of the United States of America. June 2011, 108 (23): 9402–7. PMC 3111307
 . PMID 21606324. doi:10.1073/pnas.1101026108.
. PMID 21606324. doi:10.1073/pnas.1101026108.
- ^ Teranishi, M., Nakamura, K., Morioka, H.,Yamamoto, K. and Hidema, J. The native cyclobutane pyrimidine dimer photolyase of rice is phosphorylated. Plant Physiology. 2008, 146 (4): 1941–1951. PMC 2287361
 . PMID 18235036. doi:10.1104/pp.107.110189.
. PMID 18235036. doi:10.1104/pp.107.110189.
- ^ 4.0 4.1 Selby CP, Sancar A. A cryptochrome/photolyase class of enzymes with single-stranded DNA-specific photolyase activity. Proceedings of the National Academy of Sciences of the United States of America. November 2006, 103 (47): 17696–700. PMC 1621107
 . PMID 17062752. doi:10.1073/pnas.0607993103.
. PMID 17062752. doi:10.1073/pnas.0607993103.
- ^ Lucas-Lledó JI, Lynch M. Evolution of mutation rates: phylogenomic analysis of the photolyase/cryptochrome family. Molecular Biology and Evolution. May 2009, 26 (5): 1143–53. PMC 2668831
 . PMID 19228922. doi:10.1093/molbev/msp029.
. PMID 19228922. doi:10.1093/molbev/msp029.
- ^ Garrett RH, Grisham CM. Biochemistry. Brooks/Cole, Cengage Learning. 2010. ISBN 978-0-495-10935-8. OCLC 984382855.
- ^ Sancar A. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chemical Reviews. June 2003, 103 (6): 2203–37. PMID 12797829. doi:10.1021/cr0204348.
- ^ Kulms D, Pöppelmann B, Yarosh D, Luger TA, Krutmann J, Schwarz T. Nuclear and cell membrane effects contribute independently to the induction of apoptosis in human cells exposed to UVB radiation. Proceedings of the National Academy of Sciences of the United States of America. July 1999, 96 (14): 7974–9. PMC 22172
 . PMID 10393932. doi:10.1073/pnas.96.14.7974.
. PMID 10393932. doi:10.1073/pnas.96.14.7974.
