用户:Kaguya-Taketori/RNA


核糖核酸(英语:Ribonucleic acid),简称RNA,系一类由核糖核苷酸通过3',5'-磷酸二酯键聚合而成的线性大分子[1]。自然界中的RNA通常是单链的,且RNA中最基本的四种碱基为A(腺嘌呤)、U(尿嘧啶)、G(鸟嘌呤)、C(胞嘧啶)[注 1],相对的,与RNA同为核酸的DNA通常是双链分子,且含有的含氮碱基为A(腺嘌呤)、T(胸腺嘧啶)、G(鸟嘌呤)、C(胞嘧啶)四种。
RNA有着多种多样的功能,可在遗传编码、翻译、调控、基因表达等过程中发挥作用。按RNA的功能,可将RNA分为多种类型。比如,在细胞生物中,mRNA(信使RNA)为遗传信息的传递者,它能够指导蛋白质的合成。因为mRNA有编码蛋白质的能力,它又被称为编码RNA。而其他没有编码蛋白质能力的RNA则被称为非编码RNA(ncRNA)。它们或通过催化生化反应,或通过调控或参与基因表达过程发挥相应的生物学功能。比如,tRNA(转运RNA)在翻译过程中起转运RNA的作用,rRNA(核糖体RNA)于翻译过程中起催化肽链形成的作用,sRNA(小RNA)起到调控基因表达的作用。此外,RNA病毒甚至以RNA作为它们的遗传物质。
RNA通常由DNA通过转录生成。RNA在细胞中广泛分布,真核生物的细胞核、细胞质、线粒体中都有RNA[2]:36。
与DNA的比较
[编辑]
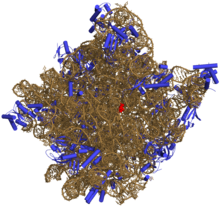
与DNA相似,大部分有生物活性的RNA,包括mRNA、tRNA、rRNA、snRNA,以及其他一些非编码RNA,虽然是单链,但含有自我互补的序列,能使得它们能进行折叠[7],形成互补双链接构(茎)。对RNA的分析表明,它们有着相对更复杂的结构。和DNA不同,RNA的二级结构并不是单纯的双螺旋,而由一系列短的二级结构构成。通过这些短的二级结构的组合,RNA甚至可以拥有与蛋白质相似的结构,并像酶那样催化化学反应(这样的RNA被称为核酶)[8]。比如,对核糖体进行分析表明,其催化成肽反应的活性位点完全由RNA构成[9]。
结构
[编辑]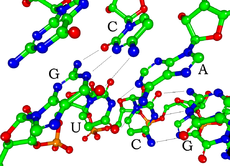
RNA的单体为核糖核苷酸,其中的戊糖为核糖,依系统命名法可将其中的碳原子从1'编号至5'。含氮碱基与1'碳原子相连。RNA中最基本的四种碱基分别为A(腺嘌呤)、U(尿嘧啶)、G(鸟嘌呤)、C(胞嘧啶)。其中,腺嘌呤和鸟嘌呤为双环的嘌呤,尿嘧啶和胞嘧啶为单环的嘧啶。磷酸基团与一个核糖残基的3'碳原子相连,与下一个核糖核苷酸的5'碳原子相连。磷酸基团在生理pH下,并不都能带上负电荷,因而RNA在生理条件下是带电荷分子(聚阴离子)。C和G、U和A、G和U之间能够形成氢键[10]。不过,碱基之间也可能发生其他一些相互作用。比如,在一个凸出部分中,一群腺嘌呤可以互相连接[11],GNRA四环中有一个G-A碱基对[10]。
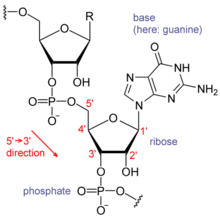
核糖的2位碳上连有羟基为RNA的一个重要结构特点。这类羟基使得RNA双链的结构应与A型构象最接近[12],不过,在单链的某些二核苷酸环境下,也有极小的可能形成DNA最常见的B型螺旋构象[13]。A型构象使得RNA双链的大沟狭窄而深,小沟浅而宽[14]。在RNA分子的构象高度可变区域(即不生成双链接构的区域),2'-OH还能攻击附近的磷酸二酯键,使得核糖-磷酸链断裂[15]。
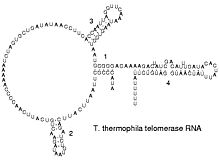
通过转录,仅仅能使RNA链上带A、U、G、C四种含氮碱基[16]不过,转录后修饰能够通过多种途径对RNA进行改造。比如,转录后修饰能够将稀有碱基假尿嘧啶(Ψ)加到RNA链上。假尿嘧啶与核糖之间的化学键是C-C键而不是尿嘧啶(U)的C-N键。胸腺嘧啶加到RNA链上的情形也很常见(最典型的例子是tRNA的TΨC环)[17]。另外,次黄嘌呤也是一种常见的稀有碱基。次黄嘌呤为腺嘌呤的脱氨产物,含有次黄嘌呤的核苷被称为肌苷(I)。在基因编码的摆动假说中,肌苷有重要的作用[18]。
除以上列出的之外,经过编辑的核苷还有100多种[19]。由修饰引发的结构性变化在tRNA中最为明显[20],假尿嘧啶与经常在rRNA中出现2'-甲氧基核糖是最常见的修饰产物[21]。这些修饰的具体作用还没有完全阐明。不过,值得注意的是,在rRNA中,许多的转录后修饰发生在高度功能化的区域,比如肽基转移酶催化中心以及亚基结合部位,似乎说明转录后修饰对RNA发挥正常功能来说相当重要[22]。
具有催化功能的单链RNA分子,和蛋白质相类似,需要特殊的RNA三级结构。通过分子内氢键形成的二级结构原件构成了三级结构的框架。二级结构形成了许多可识别的“结构域”——比如茎环结构、膨大结构(bulges)、内环结构[23]。因为RNA分子带电荷,不少二级结构和三级结构需要Mg2+等金属离子来进行稳定[24]。
自然界中的RNA均是由D-核糖核苷酸聚合而成的D-RNA。使用L-核糖核苷酸则可合成L-RNA。L-RNA对RNA酶的耐受力要强得多[25]。
合成与修饰
[编辑]RNA的合成一般由RNA聚合酶催化。RNA聚合酶以DNA为模板,通过转录合成RNA。转录起始于RNA聚合酶与启动子的结合(启动子一般位于基因的上游)。因为RNA聚合酶自带解旋酶活性,仅依靠RNA聚合酶即可实现DNA双链的解开。转录过程中,RNA聚合酶以3'端至5'端的方向读取DNA模板链,并以5'端到3'端的方向合成与之反向平行互补的RNA链。转录的终止由终止子介导。原核生物的终止子有两类:简单终止子与ρ因子依赖性终止子。简单终止子仅靠RNA形成二级结构即可终止转录,而后者在ρ因子的作用下才可以使转录终止。真核生物的转录终止则与转录后修饰密切相关[2]:323[26][27]:531-535。
在真核生物中,RNA的初始转录物通常会经过转录后修饰。比如,真核生物的mRNA大都会被加上Poly(A)尾(多腺嘌呤尾巴)以及5'端帽,mRNA前体中含有的内含子序列也会被剪接体切除[27]:580-581。
一些RNA是由RNA复制酶(RNA依赖性RNA聚合酶)以RNA为模板催化合成的。比方说,RNA病毒通过RNA复制酶复制其遗传物质[28]。另外,RNA复制酶亦参与了众多生物体的RNA干涉过程[29]。
RNA的种类
[编辑]总览
[编辑]
mRNA为携带遗传信息的RNA,它所携带的遗传信息由核糖体读取,并指导蛋白质合成。mRNA的序列决定了转译出的多肽链的氨基酸序列[30]。然而,绝大部分的RNA并不编码蛋白质(真核生物有大约97%的转录产物不编码蛋白质[31][32][33][34])。这样的RNA被称为非编码RNA(ncRNA)。它们可能由相应的基因编码(这样的基因叫RNA基因),也可以由mRNA的内含子演化而成[35]。最常见的非编码RNA当属tRNA和rRNA,它们都参与转译过程[6]。非编码RNA亦参与基因表达调控(比如转录后修饰等过程)。也有一些RNA具有催化化学反应的作用,比如对RNA进行剪接[36]、催化多肽链的合成[9],这类RNA即为核酶。
按长度分
[编辑]根据RNA链的长度,可将RNA分为小RNA(sRNA)和长链RNA(long RNA)[37]。一般地,小RNA是指长度小于200nt的RNA,长链RNA是指长度大于200nt的RNA[38]。长链RNA,也叫大RNA(large RNA),主要包括长链非编码RNA(lncRNA)和mRNA。小RNA主要包括5.8S rRNA、5S rRNA、tRNA、miRNA(微RNA)、siRNA(小干扰RNA)、snoRNA(小核仁RNA)、snRNA(小核RNA)、scRNA(细胞质小RNA)[2]:49-50。
在转译过程中
[编辑]mRNA携带遗传信息,核糖体会读取其所携带的遗传信息,并合成相应的蛋白质。核糖体以三联密码子的形式读取mRNA上的遗传信息,每一个三联密码子对应一个多肽链的氨基酸(终止密码子除外)。在真核细胞中,转录出的mRNA前体需要经过加工才能成为成熟的mRNA。加工过程会切除掉mRNA前体的内含子区(不编码的区域)。mRNA随后会从细胞核中转移到细胞质中,并与核糖体结合,在tRNA的帮助下,由核糖体读取其遗传信息并合成蛋白质。原核生物无成形细胞核,拟核和细胞质无明确界限,在转录还未完成时,核糖体就已经与mRNA结合,并开始转译过程,而且转录完成后,mRNA就会很快被核酸酶攻击降解,降解时转译过程往往还没有停止[30]。
tRNA长约80nt,携带氨基酸至核糖体上,为肽链的延伸提供底物。一个tRNA有氨基酸结合区域(5' CCA 3')以及可以与对应密码子互补的反密码子区[35]。
核糖体RNA构成了核糖体的催化核心。真核生物的核糖体包含18S、5.8S、28S以及5S rRNA(原核生物的核糖体则包含16S、5S、23S rRNA)。真核生物的三种rRNA在核仁中合成,另外一种则在核基质中合成。在细胞质中,rRNA和蛋白质装配形成一种核糖核蛋白复合物,即核糖体。核糖体与mRNA结合,并催化蛋白质的合成,一条mRNA链可同时与多个核糖体结合[30]。典型真核生物中的RNA几乎都是rRNA。
tmRNA(信使—转运RNA)在许多细菌和色素体中都有分布。它标记由缺少终止密码子的mRNA编码的蛋白质,使其降解。同时,它还能阻止核糖体的停止[39]。
Regulatory RNAs
[编辑]Several types of RNA can downregulate gene expression by being complementary to a part of an mRNA or a gene's DNA.[40][41] MicroRNAs (miRNA; 21-22 nt) are found in eukaryotes and act through RNA interference (RNAi), where an effector complex of miRNA and enzymes can cleave complementary mRNA, block the mRNA from being translated, or accelerate its degradation.[42][43]
While small interfering RNAs (siRNA; 20-25 nt) are often produced by breakdown of viral RNA, there are also endogenous sources of siRNAs.[44][45] siRNAs act through RNA interference in a fashion similar to miRNAs. Some miRNAs and siRNAs can cause genes they target to be methylated, thereby decreasing or increasing transcription of those genes.[46][47][48] Animals have Piwi-interacting RNAs (piRNA; 29-30 nt) that are active in germline cells and are thought to be a defense against transposons and play a role in gametogenesis.[49][50]
Many prokaryotes have CRISPR RNAs, a regulatory system similar to RNA interference.[51] Antisense RNAs are widespread; most downregulate a gene, but a few are activators of transcription.[52] One way antisense RNA can act is by binding to an mRNA, forming double-stranded RNA that is enzymatically degraded.[53] There are many long noncoding RNAs that regulate genes in eukaryotes,[54] one such RNA is Xist, which coats one X chromosome in female mammals and inactivates it.[55]
An mRNA may contain regulatory elements itself, such as riboswitches, in the 5' untranslated region or 3' untranslated region; these cis-regulatory elements regulate the activity of that mRNA.[56] The untranslated regions can also contain elements that regulate other genes.[57]
In RNA processing
[编辑]
Many RNAs are involved in modifying other RNAs. Introns are spliced out of pre-mRNA by spliceosomes, which contain several small nuclear RNAs (snRNA),[6] or the introns can be ribozymes that are spliced by themselves.[58] RNA can also be altered by having its nucleotides modified to other nucleotides than A, C, G and U. In eukaryotes, modifications of RNA nucleotides are in general directed by small nucleolar RNAs (snoRNA; 60-300 nt),[35] found in the nucleolus and cajal bodies. snoRNAs associate with enzymes and guide them to a spot on an RNA by basepairing to that RNA. These enzymes then perform the nucleotide modification. rRNAs and tRNAs are extensively modified, but snRNAs and mRNAs can also be the target of base modification.[59][60] RNA can also be methylated.[61][62]
RNA genomes
[编辑]Like DNA, RNA can carry genetic information. RNA viruses have genomes composed of RNA that encodes a number of proteins. The viral genome is replicated by some of those proteins, while other proteins protect the genome as the virus particle moves to a new host cell. Viroids are another group of pathogens, but they consist only of RNA, do not encode any protein and are replicated by a host plant cell's polymerase.[63]
In reverse transcription
[编辑]Reverse transcribing viruses replicate their genomes by reverse transcribing DNA copies from their RNA; these DNA copies are then transcribed to new RNA. Retrotransposons also spread by copying DNA and RNA from one another,[64] and telomerase contains an RNA that is used as template for building the ends of eukaryotic chromosomes.[65]
Double-stranded RNA
[编辑]Double-stranded RNA (dsRNA) is RNA with two complementary strands, similar to the DNA found in all cells. dsRNA forms the genetic material of some viruses (double-stranded RNA viruses). Double-stranded RNA such as viral RNA or siRNA can trigger RNA interference in eukaryotes, as well as interferon response in vertebrates.[66][67][68][69]
RNA生物学中的重大发现
[编辑]
对RNA的研究催生了许多重大的生物学发现,诺贝尔奖亦数次垂青对RNA进行研究的科学家。1868年,弗雷德里希·米歇尔发现了核酸。因为他是在细胞核中发现这种物质的,所以,他把这种物质称为“核素”(nuclein)[70]。后来,人们发现没有成形细胞核的原核细胞仍然含有核酸。早在1939年,就有人提出RNA可能参与蛋白质合成[71]。塞韦罗·奥乔亚因发现一种能够在实验室中合成RNA的酶而与阿瑟·科恩伯格获得1959年诺贝尔生理医学奖[72]。然而,由奥乔亚发现的酶(多核苷酸磷酸化酶)随后却被证明是负责RNA降解,而不是RNA合成的。1956年,亚历山大·里奇和大卫·戴维斯使两条RNA进行杂交,并制得了第一批RNA晶体。该晶体的结构可通过X射线衍射确定[73]。
1965年,罗伯特·W·霍利测定了长77nt(nt:核苷酸)的酵母菌tRNA的序列[74],这使得他与哈尔·葛宾·科拉纳、马歇尔·沃伦·尼伦伯格获得了1968年的诺贝尔生理医学奖。1967年,卡尔·乌斯提出可能存在具有催化功能的RNA。他还认为,早期的生命(自我复制的分子)可能仅用RNA来进行催化和携带遗传信息(即RNA世界假说)[75][76]。
1970年代早期,发现了逆转录病毒以及逆转录酶,证明酶可以用RNA为模板合成DNA(这和通常的遗传信息传递方向相反)。戴维·巴尔的摩、罗纳托·杜尔贝科、霍华德·马丁·特明因此获得了1975年诺贝尔生理医学奖。1976年,瓦尔特·费尔斯与他的研究团队发现了第一段与RNA病毒MS2噬菌体基因组互补的核苷酸序列[77]。
1977年,在哺乳动物病毒以及细胞基因中发现了内含子以及RNA剪接现象。这一发现使得理察·罗伯茨与菲利普·夏普获得1993年诺贝尔生理医学奖。核酶最早于1980年代早期发现,使西德尼·奥特曼、托马斯·切赫两人获1989年诺贝尔化学奖。1990年,对矮牵牛花的实验表明,引入的基因可以沉默植物体原有的基因,这一现象即RNA干扰[78][79]。
1990年左右,发现一种22nt长的RNA在秀丽隐杆线虫的发育过程中发挥作用,这种RNA即微RNA(microRNA)[80]。对RNA干扰的研究使得安德鲁·法厄与克雷格·梅洛获得2006年生理医学奖。有趣的是,当年的诺贝尔化学奖亦颁给了对RNA转录进行研究的罗杰·科恩伯格。基因调控RNA的发现使得人们开始研发以RNA为化学本质(比如siRNA(小干扰RNA))的药物来使得基因沉默[81]。
See also
[编辑]- Biomolecular structure
- DNA, RNA and proteins: The three essential macromolecules of life
- DNA
- History of RNA Biology
- List of RNA Biologists
注释
[编辑]参考
[编辑]- ^ David Hames & Nigel Hooper. Instant Notes in Biochemistry (導讀本) 3rd edition. 科学出版社. 2005: 193. ISBN 978-7-03-025218-0.
- ^ 2.0 2.1 2.2 查锡良 药立波. 生物化學與分子生物學 第八版. 人民卫生出版社. 2013. ISBN 978-7-117-17214-1.
- ^ RNA: The Versatile Molecule. University of Utah. 2015.
- ^ Nucleotides and Nucleic Acids (PDF). University of California, Los Angeles.
- ^ R.N. Shukla. Analysis of Chromosomes. ISBN 9789384568177.
- ^ 6.0 6.1 6.2 Berg JM; Tymoczko JL; Stryer L. Biochemistry 5th. WH Freeman and Company. 2002: 118–19, 781–808. ISBN 0-7167-4684-0. OCLC 179705944.
- ^ I. Tinoco & C. Bustamante. How RNA folds. J. Mol. Biol. 1999, 293 (2): 271–281. PMID 10550208. doi:10.1006/jmbi.1999.3001.
- ^ Higgs PG. RNA secondary structure: physical and computational aspects. Quarterly Reviews of Biophysics. 2000, 33 (3): 199–253. PMID 11191843. doi:10.1017/S0033583500003620.
- ^ 9.0 9.1 Nissen P; Hansen J; Ban N; Moore PB; Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000, 289 (5481): 920–30. Bibcode:2000Sci...289..920N. PMID 10937990. doi:10.1126/science.289.5481.920.
- ^ 10.0 10.1 Lee JC; Gutell RR. Diversity of base-pair conformations and their occurrence in rRNA structure and RNA structural motifs. J. Mol. Biol. 2004, 344 (5): 1225–49. PMID 15561141. doi:10.1016/j.jmb.2004.09.072.
- ^ Barciszewski J; Frederic B; Clark C. RNA biochemistry and biotechnology. Springer. 1999: 73–87. ISBN 0-7923-5862-7. OCLC 52403776.
- ^ Salazar M; Fedoroff OY; Miller JM; Ribeiro NS; Reid BR. The DNA strand in DNAoRNA hybrid duplexes is neither B-form nor A-form in solution. Biochemistry. 1992, 32 (16): 4207–15. PMID 7682844. doi:10.1021/bi00067a007.
- ^ Sedova A; Banavali NK. RNA approaches the B-form in stacked single strand dinucleotide contexts. Biopolymers. 2016, 105 (2): 65–82. PMID 26443416. doi:10.1002/bip.22750.
- ^ Hermann T; Patel DJ. RNA bulges as architectural and recognition motifs. Structure. 2000, 8 (3): R47–R54. PMID 10745015. doi:10.1016/S0969-2126(00)00110-6.
- ^ Mikkola S; Stenman E; Nurmi K; Yousefi-Salakdeh E; Strömberg R; Lönnberg H. The mechanism of the metal ion promoted cleavage of RNA phosphodiester bonds involves a general acid catalysis by the metal aquo ion on the departure of the leaving group. Perkin transactions 2. 1999, (8): 1619–26. doi:10.1039/a903691a.
- ^ Jankowski JAZ; Polak JM. Clinical gene analysis and manipulation: Tools, techniques and troubleshooting. Cambridge University Press. 1996: 14. ISBN 0-521-47896-0. OCLC 33838261.
- ^ Yu Q; Morrow CD. Identification of critical elements in the tRNA acceptor stem and TΨC loop necessary for human immunodeficiency virus type 1 infectivity. J Virol. 2001, 75 (10): 4902–6. PMC 114245
 . PMID 11312362. doi:10.1128/JVI.75.10.4902-4906.2001.
. PMID 11312362. doi:10.1128/JVI.75.10.4902-4906.2001.
- ^ Elliott MS; Trewyn RW. Inosine biosynthesis in transfer RNA by an enzymatic insertion of hypoxanthine. J. Biol. Chem. 1983, 259 (4): 2407–10. PMID 6365911.
- ^ Cantara, WA; Crain, PF; Rozenski, J; McCloskey, JA; Harris, KA; Zhang, X; Vendeix, FA; Fabris, D; Agris, PF. The RNA Modification Database, RNAMDB: 2011 update. Nucleic Acids Research. January 2011, 39 (Database issue): D195–201. PMC 3013656
 . PMID 21071406. doi:10.1093/nar/gkq1028.
. PMID 21071406. doi:10.1093/nar/gkq1028.
- ^ Söll D; RajBhandary U. TRNA: Structure, biosynthesis, and function. ASM Press. 1995: 165. ISBN 1-55581-073-X. OCLC 183036381.
- ^ Kiss T. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. The EMBO Journal. 2001, 20 (14): 3617–22. PMC 125535
 . PMID 11447102. doi:10.1093/emboj/20.14.3617.
. PMID 11447102. doi:10.1093/emboj/20.14.3617.
- ^ King TH; Liu B; McCully RR; Fournier MJ. Ribosome structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center. Molecular Cell. 2002, 11 (2): 425–35. PMID 12620230. doi:10.1016/S1097-2765(03)00040-6.
- ^ Mathews DH; Disney MD; Childs JL; Schroeder SJ; Zuker M; Turner DH. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl. Acad. Sci. USA. 2004, 101 (19): 7287–92. Bibcode:2004PNAS..101.7287M. PMC 409911
 . PMID 15123812. doi:10.1073/pnas.0401799101.
. PMID 15123812. doi:10.1073/pnas.0401799101.
- ^ Tan ZJ; Chen SJ. Salt dependence of nucleic acid hairpin stability. Biophys. J. 2008, 95 (2): 738–52. Bibcode:2008BpJ....95..738T. PMC 2440479
 . PMID 18424500. doi:10.1529/biophysj.108.131524.
. PMID 18424500. doi:10.1529/biophysj.108.131524.
- ^ Vater A; Klussmann S. Turning mirror-image oligonucleotides into drugs: the evolution of Spiegelmer therapeutics. Drug Discovery Today. January 2015, 20 (1): 147–155. PMID 25236655. doi:10.1016/j.drudis.2014.09.004.
- ^ Nudler E; Gottesman ME. Transcription termination and anti-termination in E. coli. Genes to Cells. 2002, 7 (8): 755–68. PMID 12167155. doi:10.1046/j.1365-2443.2002.00563.x.
- ^ 27.0 27.1 Jocelyn E.KREBS; et al. Gene XI. JONES&BARTLETT LEARNING(高等教育出版社出版). 2014. ISBN 978-7-04-039649-2.
- ^ Jeffrey L Hansen; Alexander M Long; Steve C Schultz. Structure of the RNA-dependent RNA polymerase of poliovirus. Structure. 1997, 5 (8): 1109–22. PMID 9309225. doi:10.1016/S0969-2126(97)00261-X.
- ^ Ahlquist P. RNA-Dependent RNA Polymerases, Viruses, and RNA Silencing. Science. 2002, 296 (5571): 1270–73. Bibcode:2002Sci...296.1270A. PMID 12016304. doi:10.1126/science.1069132.
- ^ 30.0 30.1 30.2 Cooper GC; Hausman RE. The Cell: A Molecular Approach 3rd. Sinauer. 2004: 261–76, 297, 339–44. ISBN 0-87893-214-3. OCLC 174924833.
- ^ Mattick JS; Gagen MJ. The evolution of controlled multitasked gene networks: the role of introns and other noncoding RNAs in the development of complex organisms. Mol. Biol. Evol. 1 September 2001, 18 (9): 1611–30. PMID 11504843. doi:10.1093/oxfordjournals.molbev.a003951.
- ^ Mattick, JS. Noncoding RNAs: the architects of eukaryotic complexity. EMBO Reports. 2001, 2 (11): 986–91. PMC 1084129
 . PMID 11713189. doi:10.1093/embo-reports/kve230.
. PMID 11713189. doi:10.1093/embo-reports/kve230.
- ^ Mattick JS. Challenging the dogma: the hidden layer of non-protein-coding RNAs in complex organisms (PDF). BioEssays. October 2003, 25 (10): 930–9. PMID 14505360. doi:10.1002/bies.10332.
- ^ Mattick JS. The hidden genetic program of complex organisms. Scientific American. October 2004, 291 (4): 60–7. PMID 15487671. doi:10.1038/scientificamerican1004-60. (原始内容存档于February 8, 2015).
- ^ 35.0 35.1 35.2 Wirta W. Mining the transcriptome – methods and applications. Stockholm: School of Biotechnology, Royal Institute of Technology. 2006. ISBN 91-7178-436-5. OCLC 185406288.
- ^ Rossi JJ. Ribozyme diagnostics comes of age. Chemistry & Biology. 2004, 11 (7): 894–95. PMID 15271347. doi:10.1016/j.chembiol.2004.07.002.
- ^ Storz, G. An expanding universe of noncoding RNAs.. Science. 17 May 2002, 296 (5571): 1260–3. PMID 12016301. doi:10.1126/science.1072249.
- ^ Fatica, A; Bozzoni, I. Long non-coding RNAs: new players in cell differentiation and development.. Nature Reviews Genetics. January 2014, 15 (1): 7–21. PMID 24296535. doi:10.1038/nrg3606.
- ^ Gueneau de Novoa P, Williams KP; Williams. The tmRNA website: reductive evolution of tmRNA in plastids and other endosymbionts. Nucleic Acids Res. 2004, 32 (Database issue): D104–8. PMC 308836
 . PMID 14681369. doi:10.1093/nar/gkh102.
. PMID 14681369. doi:10.1093/nar/gkh102.
- ^ Carthew, RW; Sontheimer, EJ. Origins and Mechanisms of miRNAs and siRNAs.. Cell. February 2009, 136: 642–55 [2009-02-20]. PMC 2675692
 . PMID 19239886. doi:10.1016/j.cell.2009.01.035.
. PMID 19239886. doi:10.1016/j.cell.2009.01.035.
- ^ Liang, Kung-Hao; Yeh, Chau-Ting. A gene expression restriction network mediated by sense and antisense Alu sequences located on protein-coding messenger RNAs.. BMC Genomics. 2013, 14: 325 [2013-05-11]. PMC 3655826
 . PMID 23663499. doi:10.1186/1471-2164-14-325.
. PMID 23663499. doi:10.1186/1471-2164-14-325.
- ^ Wu L; Belasco JG. Let me count the ways: mechanisms of gene regulation by miRNAs and siRNAs. Mol. Cell. January 2008, 29 (1): 1–7. PMID 18206964. doi:10.1016/j.molcel.2007.12.010.
- ^ Matzke MA; Matzke AJM. Planting the seeds of a new paradigm. PLoS Biology. 2004, 2 (5): e133. PMC 406394
 . PMID 15138502. doi:10.1371/journal.pbio.0020133.
. PMID 15138502. doi:10.1371/journal.pbio.0020133.
- ^ Vazquez F; Vaucheret H; Rajagopalan R; Lepers C; Gasciolli V; Mallory AC; Hilbert J; Bartel DP; Crété P. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Molecular Cell. 2004, 16 (1): 69–79. PMID 15469823. doi:10.1016/j.molcel.2004.09.028.
- ^ Watanabe T, Totoki Y, Toyoda A, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. May 2008, 453 (7194): 539–43. Bibcode:2008Natur.453..539W. PMID 18404146. doi:10.1038/nature06908.
- ^ Sontheimer EJ; Carthew RW. Silence from within: endogenous siRNAs and miRNAs. Cell. July 2005, 122 (1): 9–12. PMID 16009127. doi:10.1016/j.cell.2005.06.030.
- ^ Doran G. RNAi – Is one suffix sufficient?. Journal of RNAi and Gene Silencing. 2007, 3 (1): 217–19.
- ^ Pushparaj PN; Aarthi JJ; Kumar SD; Manikandan J. RNAi and RNAa — The Yin and Yang of RNAome. Bioinformation. 2008, 2 (6): 235–7. PMC 2258431
 . PMID 18317570. doi:10.6026/97320630002235.
. PMID 18317570. doi:10.6026/97320630002235.
- ^ Horwich MD; Li C; Matranga C; Vagin V; Farley G; Wang P; Zamore PD. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Current Biology. 2007, 17 (14): 1265–72. PMID 17604629. doi:10.1016/j.cub.2007.06.030.
- ^ Girard A; Sachidanandam R; Hannon GJ; Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006, 442 (7099): 199–202. Bibcode:2006Natur.442..199G. PMID 16751776. doi:10.1038/nature04917.
- ^ Horvath P; Barrangou R. CRISPR/Cas, the Immune System of Bacteria and Archaea. Science. 2010, 327 (5962): 167–70. Bibcode:2010Sci...327..167H. PMID 20056882. doi:10.1126/science.1179555.
- ^ Wagner EG; Altuvia S; Romby P. Antisense RNAs in bacteria and their genetic elements. Adv Genet. Advances in Genetics. 2002, 46: 361–98. ISBN 9780120176465. PMID 11931231. doi:10.1016/S0065-2660(02)46013-0.
- ^ Gilbert SF. Developmental Biology 7th. Sinauer. 2003: 101–3. ISBN 0-87893-258-5. OCLC 154656422.
- ^ Amaral PP; Mattick JS. Noncoding RNA in development. Mammalian genome : official journal of the International Mammalian Genome Society. October 2008, 19 (7–8): 454–92. PMID 18839252. doi:10.1007/s00335-008-9136-7.
- ^ Heard E; Mongelard F; Arnaud D; Chureau C; Vourc'h C; Avner P. Human XIST yeast artificial chromosome transgenes show partial X inactivation center function in mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA. 1999, 96 (12): 6841–46. Bibcode:1999PNAS...96.6841H. PMC 22003
 . PMID 10359800. doi:10.1073/pnas.96.12.6841.
. PMID 10359800. doi:10.1073/pnas.96.12.6841.
- ^ Batey RT. Structures of regulatory elements in mRNAs. Curr. Opin. Struct. Biol. 2006, 16 (3): 299–306. PMID 16707260. doi:10.1016/j.sbi.2006.05.001.
- ^ Scotto L; Assoian RK. A GC-rich domain with bifunctional effects on mRNA and protein levels: implications for control of transforming growth factor beta 1 expression. Mol. Cell. Biol. June 1993, 13 (6): 3588–97. PMC 359828
 . PMID 8497272.
. PMID 8497272.
- ^ Steitz TA; Steitz JA. A general two-metal-ion mechanism for catalytic RNA. Proc. Natl. Acad. Sci. U.S.A. 1993, 90 (14): 6498–502. Bibcode:1993PNAS...90.6498S. PMC 46959
 . PMID 8341661. doi:10.1073/pnas.90.14.6498.
. PMID 8341661. doi:10.1073/pnas.90.14.6498.
- ^ Xie J; Zhang M; Zhou T; Hua X; Tang L; Wu W. Sno/scaRNAbase: a curated database for small nucleolar RNAs and cajal body-specific RNAs. Nucleic Acids Res. 2007, 35 (Database issue): D183–7. PMC 1669756
 . PMID 17099227. doi:10.1093/nar/gkl873.
. PMID 17099227. doi:10.1093/nar/gkl873.
- ^ Omer AD; Ziesche S; Decatur WA; Fournier MJ; Dennis PP. RNA-modifying machines in archaea. Molecular Microbiology. 2003, 48 (3): 617–29. PMID 12694609. doi:10.1046/j.1365-2958.2003.03483.x.
- ^ Cavaillé J; Nicoloso M; Bachellerie JP. Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature. 1996, 383 (6602): 732–5. Bibcode:1996Natur.383..732C. PMID 8878486. doi:10.1038/383732a0.
- ^ Kiss-László Z; Henry Y; Bachellerie JP; Caizergues-Ferrer M; Kiss T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996, 85 (7): 1077–88. PMID 8674114. doi:10.1016/S0092-8674(00)81308-2.
- ^ Daròs JA; Elena SF; Flores R. Viroids: an Ariadne's thread into the RNA labyrinth. EMBO Rep. 2006, 7 (6): 593–8. PMC 1479586
 . PMID 16741503. doi:10.1038/sj.embor.7400706.
. PMID 16741503. doi:10.1038/sj.embor.7400706.
- ^ Kalendar R; Vicient CM; Peleg O; Anamthawat-Jonsson K; Bolshoy A; Schulman AH. Large retrotransposon derivatives: abundant, conserved but nonautonomous retroelements of barley and related genomes. Genetics. 2004, 166 (3): 1437–50. PMC 1470764
 . PMID 15082561. doi:10.1534/genetics.166.3.1437.
. PMID 15082561. doi:10.1534/genetics.166.3.1437.
- ^ Podlevsky JD; Bley CJ; Omana RV; Qi X; Chen JJ. The telomerase database. Nucleic Acids Res. 2008, 36 (Database issue): D339–43. PMC 2238860
 . PMID 18073191. doi:10.1093/nar/gkm700.
. PMID 18073191. doi:10.1093/nar/gkm700.
- ^ Blevins, T.; Rajeswaran, R.; Shivaprasad, PV.; Beknazariants, D.; Si-Ammour, A.; Park, HS.; Vazquez, F.; Robertson, D.; Meins, F. Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res. 2006, 34 (21): 6233–46. PMC 1669714
 . PMID 17090584. doi:10.1093/nar/gkl886.
. PMID 17090584. doi:10.1093/nar/gkl886.
- ^ Jana S; Chakraborty C; Nandi S; Deb JK. RNA interference: potential therapeutic targets. Appl. Microbiol. Biotechnol. 2004, 65 (6): 649–57. PMID 15372214. doi:10.1007/s00253-004-1732-1.
- ^ Schultz U; Kaspers B; Staeheli P. The interferon system of non-mammalian vertebrates. Dev. Comp. Immunol. 2004, 28 (5): 499–508. PMID 15062646. doi:10.1016/j.dci.2003.09.009.
- ^ Whitehead, K. A.; Dahlman, J. E.; Langer, R. S.; Anderson, D. G. Silencing or Stimulation? SiRNA Delivery and the Immune System. Annual Review of Chemical and Biomolecular Engineering. 2011, 2: 77–96. PMID 22432611. doi:10.1146/annurev-chembioeng-061010-114133.
- ^ Dahm R. Friedrich Miescher and the discovery of DNA. Developmental Biology. 2005, 278 (2): 274–88. PMID 15680349. doi:10.1016/j.ydbio.2004.11.028.
- ^ Caspersson T; Schultz J. Pentose nucleotides in the cytoplasm of growing tissues. Nature. 1939, 143 (3623): 602–3. Bibcode:1939Natur.143..602C. doi:10.1038/143602c0.
- ^ Ochoa S. Enzymatic synthesis of ribonucleic acid (PDF). Nobel Lecture. 1959.
- ^ Rich A; Davies, D. A New Two-Stranded Helical Structure: Polyadenylic Acid and Polyuridylic Acid. Journal of the American Chemical Society. 1956, 78 (14): 3548–3549. doi:10.1021/ja01595a086.
- ^ Holley RW, et al. Structure of a ribonucleic acid. Science. 1965, 147 (3664): 1462–65. Bibcode:1965Sci...147.1462H. PMID 14263761. doi:10.1126/science.147.3664.1462.
- ^ Siebert S. Common sequence structure properties and stable regions in RNA secondary structures (PDF). Dissertation, Albert-Ludwigs-Universität, Freiburg im Breisgau: 1. 2006. (原始内容 (PDF)存档于March 9, 2012).
- ^ Szathmáry E. The origin of the genetic code: amino acids as cofactors in an RNA world. Trends Genet. 1999, 15 (6): 223–9. PMID 10354582. doi:10.1016/S0168-9525(99)01730-8.
- ^ Fiers W, et al. Complete nucleotide-sequence of bacteriophage MS2-RNA: primary and secondary structure of replicase gene. Nature. 1976, 260 (5551): 500–7. Bibcode:1976Natur.260..500F. PMID 1264203. doi:10.1038/260500a0.
- ^ Napoli C; Lemieux C; Jorgensen R. Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell. 1990, 2 (4): 279–89. PMC 159885
 . PMID 12354959. doi:10.1105/tpc.2.4.279.
. PMID 12354959. doi:10.1105/tpc.2.4.279.
- ^ Dafny-Yelin M; Chung SM; Frankman EL; Tzfira T. pSAT RNA interference vectors: a modular series for multiple gene down-regulation in plants. Plant Physiol. December 2007, 145 (4): 1272–81. PMC 2151715
 . PMID 17766396. doi:10.1104/pp.107.106062.
. PMID 17766396. doi:10.1104/pp.107.106062.
- ^ Ruvkun G. Glimpses of a tiny RNA world. Science. 2001, 294 (5543): 797–99. PMID 11679654. doi:10.1126/science.1066315.
- ^ Fichou Y; Férec C. The potential of oligonucleotides for therapeutic applications. Trends in Biotechnology. 2006, 24 (12): 563–70. PMID 17045686. doi:10.1016/j.tibtech.2006.10.003.
External links
[编辑]- RNA World website Link collection (structures, sequences, tools, journals)
- Nucleic Acid Database Images of DNA, RNA and complexes.
- RNA Calculators
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
