DNA測序
DNA测序(英語:DNA sequencing)又稱DNA定序,是指分析特定DNA片段的碱基序列,也就是腺嘌呤(A)、胸腺嘧啶(T)、胞嘧啶(C)與鳥嘌呤(G)的排列方式。快速的DNA测序方法的出现极大地推动了生物学和医学的研究和发现。
在基础生物学研究中,和在众多的应用领域,如诊断,生物技术,法医生物学,生物系统学中,DNA序列知识已成为不可缺少的知识。具有现代的DNA测序技术的快速测序速度已经有助于达到测序完整的DNA序列,或多种类型的基因组测序和生命物种,包括人类基因组和其他许多动物,植物和微生物物种的完整DNA序列。
RNA測序則通常将RNA提取后,反转录为DNA后使用DNA测序的方法进行测序。目前应用最广泛的是由弗雷德里克·桑格发明的桑格測序[1]。新的测序方法,例如454生物科学的方法和焦磷酸测序法。
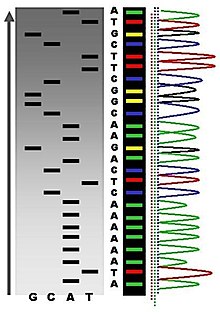
应用[编辑]
| 遗传学 |
|---|
 |
| 重要概念 |
| 历史及分支 |
| 研究 |
| 個人化醫療 |
| 精準醫學 |
DNA测序可用于确定任何生物的单个基因的序列,较大的遗传区域(即基因簇或操纵子的簇),完整的染色体或整个基因组。 DNA测序也是对RNA或蛋白质进行测序的最有效方法(通过对开放阅读框测序)。目前,DNA测序已成为生物学和其他科学领域(如医学,法医学或人类学等)的关键技术。
分子生物学[编辑]
在分子生物学中,DNA测序可被用于研究基因组及其编码的蛋白质。利用测序获得的信息,科研人员能够识别基因的变化,基因与疾病和表型的关联,并确定潜在的药物靶点。
演化生物学[编辑]
由于DNA是携带有遗传信息的大分子,在演化生物学中,DNA测序被用于研究不同生物体之间的相关性以及它们是如何演化的。
宏基因组学(或元基因组学)[编辑]
宏基因组学是一门直接取得环境中所有遗传物质的研究。环境包括但不限于水体,污水,污垢,从空气中过滤出的碎片或者从生物体采集的样本。了解在特定环境中存在哪些生物体对于生态学,流行病学,微生物学和其他领域的研究至关重要。DNA测序使研究人员能够确定微生物群中可能存在哪些类型的微生物。
医学[编辑]
医疗人员可通过对患者基因(基因组)的测序结果确定该患者是否有携带遗传性疾病的风险。需要注意的是,该方法属于基因检测,有些基因检测不会用到DNA测序技术。
法医学[编辑]
DNA测序可以与DNA图谱鉴定(基因指纹分析,英語:DNA profiling)一起用于法医鉴定和亲子鉴定。 DNA测试在过去的几十年中发展迅猛,目前已能够做到将DNA鉴定结果与被调查对象联系起来。指纹,唾液,毛囊等中的DNA特征可以将不同的生物体进行区分。测试DNA是一种可以检测DNA链中特定基因组并生成唯一的个性化DNA模型的技术。每一种有机体都有其DNA特征,并可以通过DNA测试来确定。两个人具有完全相同的DNA特征是非常罕见的,因此保证了DNA测试的成功。
历史[编辑]
DNA结构与功能的发现[编辑]

脱氧核糖核酸(DNA)最早在1869年由Friedrich Miescher发现并分离出来,但由于当时普遍认为遗传信息保存于蛋白质而不是DNA中,因此在过去几十年中DNA一直没有得到充分研究。1944年,由于Oswald Avery,Colin MacLeod和Maclyn McCarty的一些实验表明,纯化的DNA可以将一种细菌变成另一种细菌,这种情况才发生了变化。这也是首次DNA显示出改变细胞特性的能力。
1953年,James Watson和Francis Crick根据Rosalind Franklin研究的结晶X射线结构提出了他们的双螺旋DNA模型。根据该模型,DNA由彼此缠绕的两条核苷酸链组成,通过氢键连接在一起并以相反方向运行。每条链由四个互补的核苷酸组成:腺嘌呤(A),胞嘧啶(C),鸟嘌呤(G)和胸腺嘧啶(T),其中A与T配对,C与G配对。他们提出的这种结构,使得每条单链都可被用于重建另一条链,并且让遗传信息代代相传。
对蛋白质进行测序的基础首先由弗雷德里克·桑格(Frederick Sanger)的工作奠定,他于1955年完成了胰岛素(胰腺分泌的一种蛋白质)中所有氨基酸序列的测序工作。这是首个确凿的证据证明蛋白质是具有特定分子模式的化学实体,而不是悬浮在流体中的随机混合物。桑格在胰岛素测序方面的成功使得X射线晶体学家大为振奋,包括沃森和克里克,他们现在正试图理解DNA如何指导细胞内蛋白质的形成。在1954年10月弗雷德里克·桑格出席一系列讲座后不久,克里克开始发展一种理论,认为DNA中核苷酸的排列决定了蛋白质中氨基酸的序列,从而帮助确定蛋白质的功能。他于1958年发表了这一理论。
RNA测序[编辑]
RNA测序是最早的核苷酸测序形式之一。 RNA测序的主要标志是1972年和1976年Walter Fiers及其同事在根特大学(根特,比利时)确定并发表的第一个完整基因序列和噬菌体MS2的完整基因组。传统的RNA测序方法需要创建一个用于测序的互补cDNA(Complementary DNA)分子。
早期的DNA测序方法[编辑]
确定 DNA 序列的第一种方法涉及由康奈尔大学的吴瑞于1970年建立的位置特异性引物延伸策略[2]。 DNA聚合酶催化和特定核苷酸标记,这两者在当前的测序方案中都很重要,用于对λ噬菌体DNA的粘性末端进行测序[3][4][5]。在1970年至1973年间,吴瑞、R Padmanabhan及其同事证明,该方法可用于使用合成的位置特异性引物确定任何DNA序列[6][7][8]。随后弗雷德里克·桑格(Frederick Sanger)采用这种引物延伸策略在英国剑桥的英國醫學研究委員會(MRC)中心开发了更快速的DNA测序方法,并于1977年发表了“使用链终止抑制剂进行DNA测序”的方法。
全基因组测序[编辑]

第一个完整的DNA基因组测序是在1977年Φ-X174噬菌體(Phage Φ-X174)的测序工作。医学研究委员会的科学家在1984年破译了Epstein-Barr病毒的完整DNA序列,发现它含有172,282个核苷酸。 该序列的完成标志着DNA测序的一个重要转折点,它在没有病毒基因谱知识的情况下实现了DNA测序。
20世纪80年代初,Pohl及其同事开发了一种在电泳时将测序反应混合物的DNA分子转移到固定基质上的非放射性方法。随后GATC Biotech公司的DNA测序仪“Direct-Blotting-Electrophoresis-System GATC 1500”商业化,该测序仪在EU基因组测序程序的框架以及酵母酿酒酵母染色体II的完整DNA序列中广泛使用。加利福尼亚理工学院的Leroy E. Hood实验室于1986年宣布了第一台半自动DNA测序机。随后,Applied Biosystems在1987年推出了第一台全自动测序仪ABI 370。以及Dupont公司的Genesis 2000,该仪器使用了一种新的荧光标记技术,可在单一泳道中识别所有四个双脱氧核苷酸。到1990年,美国国立卫生研究院(NIH)已开始对支原体,大肠杆菌,秀丽隐杆线虫和酿酒酵母进行大规模测序实验,费用为每个碱基0.75美元。同时,人类cDNA序列的测序始于Craig Venter的实验室,试图获取人类基因组的编码部分。 1995年,Venter,Hamilton Smith及其基因组研究所(TIGR)的同事发表了第一个完整的自由生物体细菌流感嗜血杆菌(Haemophilus influenzae)的基因组。该环形染色体中含有1,830,137个碱基,其在《科学》杂志中的发表标志着全基因组鸟枪法测序的首次公开使用,摆脱了初始绘制工作的需要。
高通量測序(HTS)方法[编辑]

1990年代中後期開發了幾種新的DNA測序方法,並於 2000年在商業DNA測序儀中實施。這些方法統稱為“下一代”或“第二代”測序 (NGS) 方法,以便將它們與包括桑格测序在內的早期方法區分開來。 與第一代測序相比,NGS 技術的典型特徵是高度可擴展,允許一次對整個基因組進行測序。通常,這是通過將基因組片段化成小塊、隨機採樣片段並使用多種技術之一對其進行測序來實現的,例如下面描述的那些。 整個基因組測序是可能的,因為在一個自動化過程中同時對多個片段進行測序(命名為“大規模並行”測序)。
1990年10月26日,钱永健、Pepi Ross、Margaret Fahnestock 和 Allan J Johnston 提交了一項專利,描述了在 DNA 陣列(印跡和單個 DNA 分子)上使用可移除的 3' 阻斷劑進行逐步(“鹼基對鹼基”)測序[10]。 1996 年,斯德哥爾摩皇家理工學院的波尔·尼伦(Pål Nyrén) 和他的學生穆斯塔法·罗纳吉(Mostafa Ronaghi)發表了他們的焦磷酸測序方法[11]。
1997年4月1日,Pascal Mayer和Laurent Farinelli 向世界知識產權組織提交了描述DNA菌落測序的專利[12]。 本專利中描述的DNA樣品製備和隨機表面聚合酶链式反应 (PCR) 陣列方法,與钱永健等人的“鹼基對鹼基”測序方法相結合,現已在Illumina公司的Hi-Seq基因組測序儀中實施。
基本方法[编辑]
Maxam-Gilbert测序法[编辑]
马克萨姆-吉尔伯特测序(英语:Maxam-Gilbert sequencing)是一项由阿伦·马克萨姆与沃尔特·吉尔伯特于1976~1977年间开发的DNA测序方法。此项方法基于:对核鹼基特异性地进行局部化学改性,接下来在改性核苷酸毗邻的位点处DNA骨架发生断裂[13] 。
Sanger测序法[编辑]
Sanger(桑格)双脱氧链终止法是弗雷德里克·桑格(Frederick Sanger)于1975年发明的。测序过程需要先做一个聚合酶连锁反应(PCR)。PCR过程中,双脱氧核苷酸可能随机地被加入到正在合成中的DNA片段里。由于双脱氧核糖核苷酸又少了一个氧原子,一旦它被加入到DNA链上,这个DNA链就不能继续增加长度。最终的结果是获得所有可能获得的、不同长度的DNA片段。目前最普遍最先进的方法,是将双脱氧核糖核苷酸进行不同荧光标记。将PCR反应获得的总DNA通过毛细管电泳分离,跑到最末端的DNA就可以在激光的作用下发出荧光。由于ddATP, ddGTP, ddCTP, ddTTP(4种双脱氧核糖核苷酸)荧光标记不同,计算机可以自动根据颜色判断该位置上碱基究竟是A,T,G,C中的哪一个[14]。
高级方法和de novo测序法[编辑]
霰彈槍定序法[编辑]
霰彈槍定序法(Shotgun sequencing,又称鸟枪法)是一种广泛使用的为较长DNA测序的方法。它比傳統的定序法快速,但精確度較差。霰彈槍定序法曾經使用於塞雷拉基因組(Celera Genomics)公司所主持的人類基因組计划。
Bridge PCR[编辑]
| 此章节尚無任何内容,需要扩充。 (2021年2月3日) |
新一代测序[编辑]
随着人们对低成本测序的需求与日俱增,推动了高通量测序(high-throughput sequencing)的发展,此技术又称为二代测序、新一代测序、次世代测序;这些技术对测序过程采多路复用,同时产生上千或上百万条序列[15][16]。高通量测序技术的目的是降低DNA测序的成本,这个成本比同样可实现测序的染料终止法来得低得多[17]。超高通量测序过程中可同时运行高达500,000次的边合成边测序[18][19][20]。

| 方法 | 单分子实时测序(Pacific Bio) | 离子半导体(Ion Torrent sequencing) | 焦磷酸测序(454) | 边合成边测序(Illumina) | 边连接边测序(SOLiD sequencing) | 链终止法(Sanger sequencing) |
|---|---|---|---|---|---|---|
| 读长 | 5,500 bp to 8,500 bp avg (10,000 bp N50); maximum read length >30,000 bases[23][24][25] | up to 400 bp | 700 bp | 50 to 300 bp | 50+35 or 50+50 bp | 400 to 900 bp |
| 精确度 | 99.999% consensus accuracy; 87% single-read accuracy[26] | 98% | 99.9% | 98% | 99.9% | 99.9% |
| 每次运行可获取读段数 | 50,000 per SMRT cell, or ~400 megabases[27][28] | up to 80 million | 1 million | up to 3 billion | 1.2 to 1.4 billion | N/A |
| 每次运行耗时 | 30 minutes to 2 hours [29] | 2 hours | 24 hours | 1 to 10 days, depending upon sequencer and specified read length[30] | 1 to 2 weeks | 20 minutes to 3 hours |
| 每百万碱基所耗成本(美元) | $0.33-$1.00 | $1 | $10 | $0.05 to $0.15 | $0.13 | $2400 |
| 优势 | Longest read length. Fast. Detects 4mC, 5mC, 6mA.[31] | Less expensive equipment. Fast. | Long read size. Fast. | Potential for high sequence yield, depending upon sequencer model and desired application. | Low cost per base. | Long individual reads. Useful for many applications. |
| 劣势 | Moderate throughput. Equipment can be very expensive. | Homopolymer errors. | Runs are expensive. Homopolymer errors. | Equipment can be very expensive. Requires high concentrations of DNA. | Slower than other methods. Have issue sequencing palindromic sequence.[32] | More expensive and impractical for larger sequencing projects. |
454生物科学和焦磷酸测序法[编辑]
454测序法由454生物科学发明,是一个类似焦磷酸测序法的新方法。2003年向GenBank提交了一个腺病毒全序列[33],使得他们的技术成为Sanger测序法后第一个被用来测生物基因组全序列的新方法。454使用类似于焦磷酸测序的方法,有着相当高的读取速度,大约为5小时可以测两千万碱基对[33]。
正在开发的测序法[编辑]
纳米孔DNA测序法[编辑]
高通量测序[编辑]
高通量测序能一次对几十到几百万DNA分子进行序列测定。
參見[编辑]
参考文献[编辑]
- ^ 存档副本. [2006-11-17]. (原始内容存档于2006-11-11).
- ^ Ray Wu Faculty Profile. Cornell University. (原始内容存档于2009-03-04).
- ^ Padmanabhan R, Jay E, Wu R. Chemical synthesis of a primer and its use in the sequence analysis of the lysozyme gene of bacteriophage T4. Proceedings of the National Academy of Sciences of the United States of America. June 1974, 71 (6): 2510–4. Bibcode:1974PNAS...71.2510P. PMC 388489
 . PMID 4526223. doi:10.1073/pnas.71.6.2510
. PMID 4526223. doi:10.1073/pnas.71.6.2510  .
.
- ^ Onaga LA. Ray Wu as Fifth Business: Demonstrating Collective Memory in the History of DNA Sequencing. Studies in the History and Philosophy of Science. Part C. June 2014, 46: 1–14. PMID 24565976. doi:10.1016/j.shpsc.2013.12.006.
- ^ Wu R. Nucleotide sequence analysis of DNA. Nature New Biology. 1972, 236 (68): 198–200. PMID 4553110. doi:10.1038/newbio236198a0.
- ^ Padmanabhan R, Wu R. Nucleotide sequence analysis of DNA. IX. Use of oligonucleotides of defined sequence as primers in DNA sequence analysis. Biochem. Biophys. Res. Commun. 1972, 48 (5): 1295–302. PMID 4560009. doi:10.1016/0006-291X(72)90852-2.
- ^ Wu R, Tu CD, Padmanabhan R. Nucleotide sequence analysis of DNA. XII. The chemical synthesis and sequence analysis of a dodecadeoxynucleotide which binds to the endolysin gene of bacteriophage lambda. Biochem. Biophys. Res. Commun. 1973, 55 (4): 1092–99. PMID 4358929. doi:10.1016/S0006-291X(73)80007-5.
- ^ Jay E, Bambara R, Padmanabhan R, Wu R. DNA sequence analysis: a general, simple and rapid method for sequencing large oligodeoxyribonucleotide fragments by mapping. Nucleic Acids Research. March 1974, 1 (3): 331–53. PMC 344020
 . PMID 10793670. doi:10.1093/nar/1.3.331.
. PMID 10793670. doi:10.1093/nar/1.3.331.
- ^ Yang, Aimin; Zhang, Wei; Wang, Jiahao; Yang, Ke; Han, Yang; Zhang, Limin. Review on the Application of Machine Learning Algorithms in the Sequence Data Mining of DNA. Frontiers in Bioengineering and Biotechnology. 2020, 8: 1032. PMC 7498545
 . PMID 33015010. doi:10.3389/fbioe.2020.01032
. PMID 33015010. doi:10.3389/fbioe.2020.01032  .
.
- ^ Espacenet – Bibliographic data. worldwide.espacenet.com. [2021-12-04]. (原始内容存档于2022-01-10).
- ^ Ronaghi M, Karamohamed S, Pettersson B, Uhlén M, Nyrén P. Real-time DNA sequencing using detection of pyrophosphate release. Analytical Biochemistry. 1996, 242 (1): 84–89. PMID 8923969. doi:10.1006/abio.1996.0432.
- ^ Kawashima, Eric H.; Laurent Farinelli; Pascal Mayer. Patent: Method of nucleic acid amplification. 2005-05-12 [2012-12-22]. (原始内容存档于22 February 2013).
- ^ Maxam AM, Gilbert W. A new method for sequencing DNA. Proc. Natl. Acad. Sci. U.S.A. February 1977, 74 (2): 560–4. Bibcode:1977PNAS...74..560M. PMC 392330
 . PMID 265521. doi:10.1073/pnas.74.2.560.
. PMID 265521. doi:10.1073/pnas.74.2.560.
- ^ Sanger sequencing. 2020年3月20日 [2020年3月27日]. (原始内容存档于2020年3月29日) –通过Wikipedia.
- ^ Hall, Nell. Advanced sequencing technologies and their wider impact in microbiology. J. Exp. Biol. May 2007, 209 (Pt 9): 1518–1525. PMID 17449817. doi:10.1242/jeb.001370.

- ^ Church, George M. Genomes for all. Sci. Am. January 2006, 294 (1): 46–54. PMID 16468433. doi:10.1038/scientificamerican0106-46.

- ^ Schuster SC. Next-generation sequencing transforms today's biology. Nat. Methods. January 2008, 5 (1): 16–18. PMID 18165802. S2CID 1465786. doi:10.1038/nmeth1156.
- ^ Kalb, Gilbert; Moxley, Robert. Massively Parallel, Optical, and Neural Computing in the United States. IOS Press. 1992. ISBN 90-5199-097-9.[页码请求]
- ^ John R. ten Bosch, Wayne W. Grody. Keeping up with the next generation: massively parallel sequencing in clinical diagnostics. The Journal of molecular diagnostics: JMD. 2008-11, 10 (6): 484–492 [2019-02-12]. ISSN 1525-1578. PMC 2570630
 . PMID 18832462. doi:10.2353/jmoldx.2008.080027. (原始内容存档于2019-06-12).
. PMID 18832462. doi:10.2353/jmoldx.2008.080027. (原始内容存档于2019-06-12).
- ^ Tracy Tucker, Marco Marra, Jan M. Friedman. Massively parallel sequencing: the next big thing in genetic medicine. American Journal of Human Genetics. 2009-08, 85 (2): 142–154 [2019-02-12]. ISSN 1537-6605. PMC 2725244
 . PMID 19679224. doi:10.1016/j.ajhg.2009.06.022. (原始内容存档于2019-06-06).
. PMID 19679224. doi:10.1016/j.ajhg.2009.06.022. (原始内容存档于2019-06-06).
- ^ Quail, Michael; Smith, Miriam E; Coupland, Paul; et al. A tale of three next generation sequencing platforms: comparison of Ion torrent, pacific biosciences and illumina MiSeq sequencers. BMC Genomics. 1 January 2012, 13 (1): 341. PMC 3431227
 . PMID 22827831. doi:10.1186/1471-2164-13-341.
. PMID 22827831. doi:10.1186/1471-2164-13-341.
- ^ Liu, Lin; Li, Yinhu; Li, Siliang; et al. Comparison of Next-Generation Sequencing Systems. Journal of Biomedicine and Biotechnology (Hindawi Publishing Corporation). 1 January 2012, 2012: 1–11. doi:10.1155/2012/251364.

- ^ New Products: PacBio's RS II; Cufflinks. GenomeWeb. [2020-03-27]. (原始内容存档于2020-03-27).
- ^ After a Year of Testing, Two Early PacBio Customers Expect More Routine Use of RS Sequencer in 2012. GenomeWeb. 10 January 2012 [2014-02-08]. (原始内容存档于2013-12-12).

- ^ Inc, Pacific Biosciences of California. Pacific Biosciences Introduces New Chemistry With Longer Read Lengths to Detect Novel Features in DNA Sequence and Advance Genome Studies of Large Organisms. GlobeNewswire News Room. 2013年10月3日 [2020年3月27日]. (原始内容存档于2020年3月27日).
- ^ Chin, Chen-Shan; Alexander, David H.; Marks, Patrick; Klammer, Aaron A.; Drake, James; Heiner, Cheryl; Clum, Alicia; Copeland, Alex; Huddleston, John; Eichler, Evan E.; Turner, Stephen W.; Korlach, Jonas. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nature Methods. 2013年6月27日, 10 (6): 563–569 [2020年3月27日]. doi:10.1038/nmeth.2474. (原始内容存档于2020年3月29日) –通过www.nature.com.
- ^ De novo bacterial genome assembly: a solved problem?. 2013年7月5日 [2020年3月27日]. (原始内容存档于2020年3月27日).
- ^ Rasko, David A.; Webster, Dale R.; Sahl, Jason W.; et al. Origins of the Strain Causing an Outbreak of Hemolytic–Uremic Syndrome in Germany. N Engl J Med. 25 August 2011, 365 (8): 709–717. doi:10.1056/NEJMoa1106920.

- ^ Tran, Ben; Brown, Andrew M.K.; Bedard, Philippe L.; Winquist, Eric; Goss, Glenwood D.; Hotte, Sebastien J.; Welch, Stephen A.; Hirte, Hal W.; Zhang, Tong; Stein, Lincoln D.; Ferretti, Vincent; Watt, Stuart; Jiao, Wei; Ng, Karen; Ghai, Sangeet; Shaw, Patricia; Petrocelli, Teresa; Hudson, Thomas J.; Neel, Benjamin G.; et al. Feasibility of real time next generation sequencing of cancer genes linked to drug response: Results from a clinical trial. Int. J. Cancer. 1 January 2012: 1547–1555. doi:10.1002/ijc.27817.

- ^ van Vliet, Arnoud H.M. Next generation sequencing of microbial transcriptomes: challenges and opportunities. FEMS Microbiology Letters. 1 January 2010, 302 (1): 1–7. doi:10.1111/j.1574-6968.2009.01767.x.

- ^ Murray I. A.; Clark, T. A.; Morgan, R. D.; Boitano, M.; Anton, B. P.; Luong, K.; Fomenkov, A.; Turner, S. W.; Korlach, J.; Roberts, R. J. The methylomes of six bacteria. Nucleic Acids Research. 2 October 2012, 40 (22): 11450–62. PMC 3526280
 . PMID 23034806. doi:10.1093/nar/gks891.
. PMID 23034806. doi:10.1093/nar/gks891.
- ^ Yu-Feng Huang, Sheng-Chung Chen, Yih-Shien Chiang, Tzu-Han Chen & Kuo-Ping Chiu. Palindromic sequence impedes sequencing-by-ligation mechanism. BMC systems biology. 2012,. 6 Suppl 2: S10. PMID 23281822. doi:10.1186/1752-0509-6-S2-S10.
- ^ 33.0 33.1 About 454 - Overview. [2006-11-17]. (原始内容存档于2006-10-29).
| ||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||
|

