酶

| 生物學的一部分 |
| 生物化學 |
|---|
 |
| 關鍵部分 |
| 歷史和主題 |
| 詞彙 |
| 生物化學主題 |
酶(英語:Enzyme),又稱酵素,是一類大分子生物催化劑。酶能加快化學反應的速度(即具有催化作用)。由酶催化的反應中,反應物稱爲受質,生成的物質稱爲產物。幾乎所有細胞內的代謝過程都離不開酶。酶能大大加快這些過程中各化學反應進行的速率,使代謝產生的物質和能量能滿足生物體的需求[1]:8.1。細胞中酶的類型對可在該細胞中發生的代謝途徑的類型起決定作用。對酶進行研究的學科稱爲酶學(enzymology)。
目前已知酶可以催化超過5000種生化反應[2]。大部分酶是蛋白質,有少部分酶是具有催化活性的RNA分子,這些酶被稱為核酶。酶的特異性是由其獨特的三級結構決定的。
和所有的催化劑一樣,酶通過降低反應活化能加快化學反應的速率。一些酶可以將受質轉化爲產物的速率提高數百萬倍。一個比較極端的例子是乳清苷-5'-磷酸脫羧酶。該酶可以使在無催化劑條件下需要進行數百萬年的化學反應在幾毫秒內完成[3][4]。從化學原理上講,酶和其它所有催化劑一樣,反應不會使其物質量發生變化。酶亦不能改變化學平衡,這一點和其它催化劑也是一樣的。酶和其它催化劑的不同之處在於,它們的專一性要強得多。一些分子可以影響酶的活性。如酶抑制劑能降低酶的活性,酶活化劑能提高酶的活性。許多藥物及毒物是酶的抑制劑。當超出或小於適宜的溫度和pH值後,酶的活性會顯著下降。
酶在工業和人們的日常生活中的應用也非常廣泛。例如,藥廠用特定的合成酶來合成抗生素;洗衣粉中添加酶能加速附著在衣物上的蛋白質、澱粉或脂肪漬的分解;嫩肉粉中加入木瓜蛋白酶能將蛋白質分解爲稍小的分子,使肉的口感更嫩滑。
發現及研究史[編輯]
酶的發現來源於人們對發酵機理的逐漸了解。早在18世紀末和19世紀初,人們就認識到食物在胃中被消化,[5]用植物的提取液可以將澱粉轉化為糖,但對於其對應的機理則並不了解。[6]
十八世紀義大利的生物學家斯巴蘭贊尼在火雞和鵝的砂囊中發現可以分解碎肉和麥粒的液體,後來法國的兩位化學家沛因與&貝索茲則由大麥和水磨碎的混合物中提煉出能分解澱粉的物質。
1878年,德國生理學家威廉·屈內首次提出了酶的概念。隨後,酶被用於專指胃蛋白酶等一類非活體物質,而酵素則被用於指由活體細胞產生的催化活性。

這種對酶的錯誤認識很快得到糾正。1897年,德國科學家愛德華·比希納開始對不含細胞的酵母提取液進行發酵研究,通過在柏林洪堡大學所做的一系列實驗最終證明發酵過程並不需要完整的活細胞存在。[7]他將其中能夠發揮發酵作用的酶命名為發酵酶。[8]這一貢獻打開了通向現代酶學與現代生物化學的大門,其本人也因「發現無細胞發酵及相應的生化研究」而獲得了1907年的諾貝爾化學獎。在此之後,酶和酵素兩個概念合二為一,並依據比希納的命名方法,酶的發現者們根據其所催化的反應將它們命名。通常酶的英文名稱是在催化受質或者反應類型的名字最後加上-ase的後綴,而對應中文命名也採用類似方法,即在名字最後加上「酶」。例如,乳糖酶是能夠剪切乳糖的酶;DNA聚合酶能夠催化DNA聚合反應。
人們在認識到酶是一類不依賴於活體細胞的物質後,下一步工作就是鑑定其生化組成成分。許多早期研究者指出,一些蛋白質與酶的催化活性相關;但包括諾貝爾獎得主里夏德·維爾施泰特在內的部分科學家認為酶不是蛋白質,他們辯稱那些蛋白質只是酶分子的攜帶者,蛋白質本身並不具有催化活性。1926年,美國生物化學家詹姆斯·薩姆納完成了一個決定性的實驗。他首次從刀豆得到尿素酶結晶,並證明了尿素酶的蛋白質本質。其後,薩姆納在1931年在過氧化氫酶的研究中再次證實了酶為蛋白質。約翰·霍華德·諾思羅普和溫德爾·梅雷迪思·斯坦利通過對胃蛋白酶、胰蛋白酶和胰凝乳蛋白酶等消化性蛋白酶的研究,最終確認蛋白質可以是酶。以後陸續發現的兩千餘種酶均證明酶的化學本質是蛋白質。以上三位科學家因此獲得1946年度諾貝爾化學獎。[9]
由於蛋白質可以結晶,通過X射線晶體學就可以對酶的三維結構進行研究。第一個獲得結構解析的酶分子是溶菌酶,一種在眼淚、唾液和蛋清中含量豐富的酶,其功能是溶解細菌外殼。溶菌酶結構由大衛·菲利浦所領導的研究組解析,並於1965年發表。[10]這一成果的發表標誌著結構生物學研究的開始,高解析度的酶三維結構使得對於酶在分子水平上的工作機制的了解成為可能。
1980年代,托馬斯·切赫和雪梨·奧爾特曼分別從四膜蟲的rRNA前體的加工研究和細菌的核糖核酸酶P複合物的研究中都發現RNA本身具有自我催化作用,並提出了核酶的概念。這是第一次發現蛋白質以外的具有催化活性的生物分子。 1989年,其二人也因此獲得諾貝爾化學獎。[11]
命名規則[編輯]
酶的命名是衍生自其受質或是要催化的化學反應,在字尾會加上-ase[1]:8.1.3。例如乳糖酶、醇去氫酶及DNA聚合酶。但有些化學反應可以由幾種不同的酶催化,這些酶稱為同工酶[1]:10.3,而上述的命名法無法處理同工酶的情形。
國際生物化學與分子生物學聯盟提出了酶的命名法,也就是EC編號。每一個酶用一個四位數的數字表示,前面再加上"EC"。第一位數字是酶依酶促反應的機制來分類[12]。
依照第一位數字,可以分為以下六類:
- EC 1 氧化還原酶:催化氧化還原反應的酶類,例如乳酸去氫酶、琥珀酸去氫酶、細胞色素氧化酶、過氧化氫酶、過氧化物酶等。
- EC 2 轉移酶:轉移官能基(例如甲基或是磷酸基團)的酶類,例如甲基轉移酶、胺基轉移酶、己醣激酶、磷酸化酶等。
- EC 3 水解酶:催化受質發生水解反應的酶類,例如澱粉酶、蛋白酶、脂肪酶、磷酸酶等。
- EC 4 解離酶:用氧化及水解反應以外的方式移去基團的酶類,例如碳酸酐酶、醛縮酶、檸檬酸合酶等。
- EC 5 異構酶:催化分子同分異構反應的酶類,例如磷酸丙醣異構酶、消旋酶等。
- EC 6 連接酶:用共價鍵結合二個分子的酶類,例如麩胺醯胺合成酶、丙酮酸羧化酶等。。
上述分類會再依受質、生成物及化學結構來分類。用四位數字可以完整的描述一個酶。例如,己醣激酶(EC 2.7.1.1)是轉移酶(EC 2),會將磷酸基團(EC 7)加到六碳醣中,是一個含有醇基的分子(EC 2.7.1)[13]。
結構[編輯]
酶大都是球狀蛋白,以單體或聚成複合物對反應進行催化。和其他的蛋白質一樣,酶的三維結構是通過多肽鏈摺疊形成的。胺基酸的序列(一級結構)能決定蛋白質的三維結構,進而影響酶的催化活性[14]。儘管結構決定功能是一條具普適性的規則,一種新的酶的活性不能僅僅通過其結構預測[15]。加熱時或與化學變性劑接觸時,酶結構會發生去摺疊(即變性),原有的結構被打亂,活性也往往隨之喪失[16]。在溫度超過正常水平時,酶就會變性。因此,不難推斷生活在火山環境(比如熱泉)中的細菌的酶具有很強的耐熱性。這些酶使高溫條件下酶促反應的發生成爲可能,在工業上具有很高的利用價值。
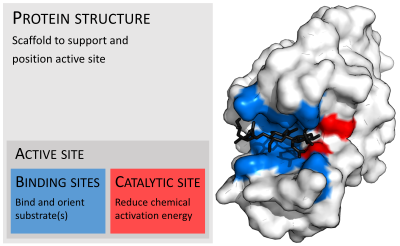
酶通常比受質大得多。酶的肽鏈長度從62個胺基酸殘基的4-草醯巴豆酸異構酶的單體[17]到長度超過2,500個胺基酸殘基的動物脂肪酸合酶[18]。酶的結構只有一小部分(大約2-4個胺基酸)是直接與催化相關的。這部分稱爲催化位點[19]。催化位點通常與一個或多個與受質結合的結合位點相連。催化位點與結合位點共同組成了酶的活性位點。酶的其餘部分起維持活性位點準確的方向以及動力學特性的作用[20]。
在一些酶中,催化與任何一個胺基酸都沒有關係。這類酶另有與催化輔助因子結合的位點[20]。一些酶亦可能包含異位位點。小分子與異位位點的結合可使酶發生構象改變,進而使酶的活性降低或升高[21]。
一些具有生物催化活性的RNA分子稱爲核酶。這類分子可能單獨發揮催化作用,也可能在與蛋白質結合成複合物的條件下發揮催化作用。最常見的核酶應是核糖體。核糖體是蛋白質以及具有催化活性的RNA的複合物。核糖體的活性位點完全由RNA組成,而蛋白質僅起支架的作用[1]:2.2[22]:695-701。
機制[編輯]
受質結合[編輯]
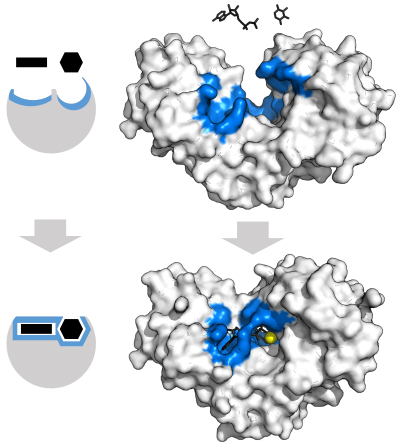
酶在催化化學反應前必須要與受質結合。酶具有很強的專一性,通常僅能與寥寥數種受質結合,催化一種或幾種反應。專一性通過結合區的形狀、電荷、疏水/親水性與受質互補實現。因此,酶可以用來區分化學選擇性上、區域選擇性上、立體專一性上有所不同的結構相似的分子[23]。
一些與基因組複製與表達相關的酶具有很高的專一性和準確性,表現在一些酶所具有的校對機制。以DNA聚合酶爲例,這類酶先催化DNA鏈的合成,再檢查新加上的鹼基是否正確[24]。校對機制確保了酶的極高準確性,哺乳動物的高保真DNA聚合酶在每一億次反應中才會出一次錯誤[1]:5.3.1。RNA聚合酶、胺醯-tRNA合成酶[25]、核糖體[26]也有與DNA聚合酶類似的校對機制[27]。
相對應地,另外一些酶則表現出多受質特異性(的現象。這類酶專一性弱,能與一系列生理上相關的受質反應。很多此類酶偶然還會出現一定程度的副反應活性,而這可能是進化出新功能酶的起點[28][29]。
「鑰匙和鎖」模型[編輯]
爲了解釋觀察到的酶的特異性,1894年,赫爾曼·埃米爾·費歇爾提出,酶和受質靠著互補的幾何形狀精準地結合在一起[30]。這一理論即通常所說的「鑰匙和鎖」模型[1]:8.3.2。這一早期的理論解釋了酶的專一性,但卻沒能解釋酶的過渡態爲何能穩定存在[31]。
誘導契合模型[編輯]
1958年,丹尼爾·科甚蘭提出了一個對鑰匙與鎖模型進行修正的理論:酶的結構相對靈活,受質與酶(活性位點)作用時,活性位點會不斷改變結構[32]。受質不是簡單地與一個剛性的活性位點結合。組成活性位點的胺基酸側鏈的準確有序排布保證酶能執行催化功能。醣苷酶等酶,當受質分子與活性位點結合時,受質分子亦會發生輕微的形狀改變[33]。直到受質與酶發生完全結合,分子形狀和電荷排佈都最終確定,活性位點都會不斷發生結構變化[34]誘導契合可以通過結構校對機制在噪音和競爭物存在的條件下增強分子識別的保真度[35]。
催化機理[編輯]
酶可以通過多種方式加快化學反應的進行速度,基本機理都是降低反應的活化能(ΔG‡,吉布斯自由能)[36]。
酶可以同時使用以上多種催化機制來催化反應。比如,胰蛋白酶先通過一個催化三聯體進行共價催化產生中間態,再藉助氧負離子洞穩定過渡態的電荷排佈,水解過程的完成則依賴有序排列的水分子受質。
動態結構[編輯]
酶的結構並不是剛性的、恆定不變的。酶的內部結構會發生複雜的動力學變化。酶的結構可能在反應過程中發生變化,單個胺基酸殘基、一個轉角、一個二級結構單位,乃至整個結構域的位置都可能發生改變。酶內部結構的變化能使差異度不大、且能發生互變的多種構象體在熱力學平衡狀態下共存,從而形成所謂構象系綜。在系綜狀態下,酶的每一結構狀態或者構象體都可能與酶的功能的某一部分有關。舉例來說,二氫葉酸還原酶的不同構象就分別和受質結合、催化、輔助因子釋放、產物釋放相關[42]。
異位調節[編輯]
別構中心是酶結構上的結合口袋,遠離活性中心,可以與細胞環境中的一些分子結合。和別構中心結合的分子往往可以改變酶的構象或是酶的動態結構,從而影響酶活性中心的反應速率[43]。因此,別構反應可以使酶被活化或被抑制。別構調節對代謝通路非常重要,代謝酶往往可以和通路上游或是下游的代謝物發生別構效應,而產生反饋調控,即根據通路其他部分上的代謝通量來調整酶的活性[44]。
輔因子[編輯]

一些酶並不需要額外的組分就能就能正常發揮作用,另外一些酶則要在和輔助因子結合後才能顯示出活性[45]。輔助因子可以是無機物(如金屬離子、鐵硫簇),也可以是有機物(比如黃素和血基質)。有機輔助因子如果在反應中會與酶分離則爲輔酶,如果與酶緊密結合則爲輔基。有機的輔基可能與酶發生共價結合(丙酮酸羧化酶與生物素之間即發生共價結合)[46]。
碳酸酐酶即是一類含有輔助因子的酶。理察森圖顯示,碳酸酐酶的肽鏈環繞在一個鋅離子(值得注意的是,鋅離子亦是活性位點的一部分)周圍,並與這個鋅離子結合在一起[47]這些與酶緊密結合的離子或分子通常位於活性位點之中,並且能夠參與催化反應[1]:8.1.1。例如,黃素以及血紅素即會參與氧化還原反應[1]:17。
需要輔助因子才可以發揮作用的酶,處於未與輔助因子狀態時稱爲「脫輔酶」(apoenzymes或apoproteins),當其與輔助因子結合後則稱爲「全酶」(holoenzyme)。不過,值得注意的是,名詞全酶亦可以指含有多個亞基的酶,如DNA聚合酶。不過,在本文中,全酶是指含有所有發揮活性所需的亞基的酶[1]:8.1.1。
輔酶[編輯]
輔酶是一類與酶結合的小分子有機物,輔酶與酶結合的強度因輔酶和酶的種類而異,或強或弱。輔酶能夠將化學基團從一個酶轉移到另一個酶上[48]。NADH、NADPH爲兩種常見的輔酶。核黃素、硫胺、葉酸等維生素類輔酶人體不能合成,需要通過膳食補充。輔酶能攜帶化學基團,如NAD或NADP能攜帶負氫離子、ATP能攜帶磷酸基團、輔酶A能攜帶乙醯基團、葉酸基團能攜帶甲醯基、次甲基,或甲基、S-腺苷甲硫胺酸能攜帶甲基[48]。
輔酶在酶促反應發生後化學結構會發生改變。因此,可以將看作一類普遍存在的特殊受質。許多酶都有與之匹配的輔酶,2015年已知大約1000種酶使用NADH作爲輔酶[49]。
輔酶通常能不斷再生,濃度能始終維持在一個恆定不變的水平上。舉例來說,NADPH能通過磷酸戊醣途徑再生,S-腺苷甲硫胺酸能通過甲硫胺酸腺苷轉移酶催化的反應生成。持續不斷的再生意味著總量不多的輔酶也能以很快的速度消耗。舉例來說,人類每天會更新重量與自身體重相等的ATP[50]。
熱力學[編輯]

與其他催化劑一樣,酶並不改變反應的平衡常數,而是通過降低反應的活化能來加快反應速率(見右圖)。通常情況下,反應在酶存在或不存在的兩種條件下,其反應方向是相同的,只是前者的反應速度更快一些。但必須指出的是,在酶不存在的情況下,受質可以通過其他不受催化的「自由」反應生成不同的產物,原因是這些不同產物的形成速度更快。
酶可以連接兩個或多個反應,因此可以用一個熱力學上更容易發生的反應去「驅動」另一個熱力學上不容易發生的反應。例如,細胞常常通過ATP被酶水解所產生的能量來驅動其他化學反應。[51]
酶可以同等地催化正向反應和逆向反應,而並不改變反應自身的化學平衡。例如,碳酸酐酶可以催化如下兩個可逆反應,催化哪一種反應則是依賴於反應物濃度。[52]
反應式中「*」表示「碳酸酐酶」
當然,如果反應平衡極大地趨向於某一方向,比如釋放高能量的反應,而逆反應不可能有效的發生,則此時酶實際上只催化熱力學上允許的方向,而不催化其逆反應。
動力學[編輯]

| 未解決的化學問題:為什麼一些酶的表現快於擴散動力學? |
酶動力學是研究酶結合受質能力和催化反應速率的科學。研究者通過酶反應分析法來獲得用於酶動力學分析的反應速率資料。
1902年,維克多·亨利提出了酶動力學的定量理論;[53]隨後該理論得到他人證實並擴展為米氏方程。[54]亨利最大貢獻在於其首次提出酶催化反應由兩步組成:首先,受質可逆地結合到酶上,形成酶-受質複合物;然後,酶完成對對應化學反應的催化,並釋放生成的產物(見左圖)。
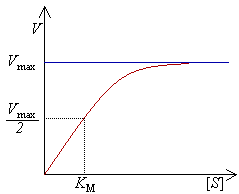
酶可以在一秒鐘內催化數百萬個反應。例如,乳清酸核苷5'-磷酸脫羧酶所催化的反應在無酶情況下,需要七千八百萬年才能將一半的受質轉化為產物;而同樣的反應過程,如果加入這種脫羧酶,則需要的時間只有25毫秒。[55]酶催化速率依賴於反應條件和受質濃度。如果反應條件中存在能夠將蛋白解鏈的因素,如高溫、極端的pH和高的鹽濃度,都會破壞酶的活性;而提高反應體系中的受質濃度則會增加酶的活性。在酶濃度固定的情況下,隨著受質濃度的不斷升高,酶催化的反應速率也不斷加快並趨向於最大反應速率(Vmax,見右圖的飽和曲線)。出現這種現象的原因是,當反應體系中受質的濃度升高,越來越多自由狀態下的酶分子結合受質形成酶-受質複合物;當所有酶分子的活性位點都被受質飽和結合,即所有酶分子形成酶-受質複合物時,催化的反應速率達到最大。當然,Vmax並不是酶唯一的動力學常數,要達到一定反應速率所需的受質濃度也是一個重要的動力學指標。這一動力學指標即米氏常數(Km),指的是達到Vmax值一半的反應速率所需的受質濃度(見右圖)。對於特定的受質,每一種酶都有其特徵Km值,表示受質與酶之間的結合強度(Km值越低,結合越牢固,親和力越高)。另一個重要的動力學指標是kcat,定義為一個酶活性位點在一秒鐘內催化受質的數量,用於表示酶催化特定受質的能力。
酶的催化效率可以用kcat/Km來衡量。這一表示式又被稱為特異性常數,其包含了催化反應中所有步驟的反應常數。由於特異性常數同時反映了酶對受質的親和力和催化能力,因此可以用於比較不同酶對於特定受質的 催化效率或同一種酶對於不同受質的催化效率。特異性常數的理論最大值,又稱為擴散極限,約為108至109 M−1s−1;此時,酶與受質的每一次碰撞都會導致受質被催化,因此產物的生成速率不再為反應速率所主導,而分子的擴散速率起到了決定性作用。酶的這種特性被稱為「催化完美性」或「動力學完美性」。相關的酶的例子有磷酸丙醣異構酶、碳酸酐酶、乙醯膽鹼酯酶、過氧化氫酶、延胡索酸酶、β-內醯胺酶和超氧化物歧化酶。
米氏方程是基於質量作用定律而確立的,而該定律則基於自由擴散和熱動力學驅動的碰撞這些假定。然而,由於酶/受質/產物的高濃度和相分離或者一維/二維分子運動,許多生化或細胞進程明顯偏離質量作用定律的假定。[56]在這些情況下,可以應用分形米氏方程。[57][58][59][60]
存在一些酶,它們的催化產物動力學速率甚至高於分子擴散速率,這種現象無法用目前公認的理論來解釋。有多種理論模型被提出來解釋這類現象。其中,部分情況可以用酶對受質的附加效應來解釋,即一些酶被認為可以通過雙偶極電場來捕捉受質以及將受質以正確方位擺放到催化活性位點。另一種理論模型引入了基於量子理論的穿隧效應,即質子或電子可以穿過活化能壘(就如同穿過隧道一般),但關於穿隧效應還有較多爭議。[61][62]有報導發現色胺中質子存在量子穿隧效應。[63]因此,有研究者相信在酶催化中也存在著穿隧效應,可以直接穿過反應能壘,而不是像傳統理論模型的方式通過降低能壘達到催化效果。有相關的實驗報導提出在一種醇去氫酶的催化反應中存在穿隧效應,[64]但穿隧效應是否在酶催化反應中普遍存在並未有定論。[65]
抑制作用[編輯]
酶的催化活性可以被多種抑制劑所降低。[66]

可逆抑制作用[編輯]
可逆抑制作用的類型有多種,它們的共同特點在於抑制劑對酶活性的抑制反應具有可逆性。
競爭性抑制作用[編輯]
抑制劑與受質競爭結合酶的活性位點(抑制劑和受質不能同時結合到活性位點),也就意味著它們不能同時結合到酶上。[68]對於競爭性抑制作用,催化反應的最大反應速率值沒有變,但是需要更高的受質濃度,反映在表觀Km值的增加。
非競爭性抑制作用[編輯]
非競爭性抑制抑制劑可以與受質同時結合到酶上,即抑制劑不結合到活性位點。酶-抑制劑複合物(EI)或酶-抑制劑-受質複合物(EIS)都沒有催化活性。與競爭性抑制作用相比,非競爭性抑制作用不能通過提高受質濃度來達到所需反應速度,即表觀最大反應速率Vmax的值變小;而同時,由於抑制劑不影響受質與酶的結合,因此Km值保持不變。
反競爭性抑制作用[編輯]
反競爭性抑制作用比較少見:抑制劑不能與處於自由狀態下的酶結合,而只能和酶-受質複合物(ES)結合,在酶反應動力學上表現為Vmax和Km值都變小。這種抑制作用可能發生在多亞基酶中。
複合抑制作用[編輯]
這種抑制作用與非競爭性抑制作用比較相似,區別在於EIS複合物殘留有部分酶的活性。在許多生物體中,這類抑制劑可以作為負反饋機制的組成部分。若一個酶體系生產了過多的產物,那麼產物就會抑制合成該產物的酶體系中第一個酶的活性,這就可以保證一旦合成足夠多的產物後,該產物的合成速率會下降或停止。受這種抑制作用調控的酶通常為多亞基酶,並具有與調控產物結合的異位結合位點。這種抑制作用的反應速率與受質濃度的關係圖不再是雙曲線形而是S形。
不可逆抑制作用[編輯]
不可逆抑制劑可以與酶結合形成共價連接,而其他抑制作用中酶與抑制劑之間都是非共價結合。這種抑制作用是不可逆的,酶一旦被抑制後就無法再恢復活性狀態。這類抑制劑包括二氟甲基鳥胺酸(一種可用於治療寄生蟲導致的昏睡症的藥物[69])、苯甲基磺醯氟(PMSF)、青黴素和阿司匹林。這些藥物都是與酶活性位點結合併被活化,然後與活性位點處的一個或多個胺基酸殘基發生不可逆的反應形成共價連接。
抑制劑的用途[編輯]
酶抑制劑常被用作藥物,同樣也可以被作為毒藥使用。而藥物和毒藥之間的差別通常非常小,大多數的藥物都有一定程度的毒性,正如帕拉塞爾蘇斯所言:「所有東西都有毒,沒有什麼是無毒的」(「In all things there is a poison, and there is nothing without a poison」)。[70]相同的,抗生素和其他抗感染藥物只是特異性地對病原體而不是對宿主有毒性。
一個獲得廣泛應用的抑制劑藥物是阿司匹林,它可以抑制環加氧酶的活性,而環加氧酶可以生產炎症反應信使前列腺素,因此,阿司匹林可以起到抑制疼痛與炎症的作用。而劇毒毒藥氰化物可以通過結合細胞色素氧化酶位點處的銅和鐵原子不可逆地抑制酶活性,從而抑制細胞的呼吸作用。[71]
生物學功能[編輯]
在生物體內,酶發揮著非常廣泛的功能。訊息傳遞和細胞活動的調控都離不開酶,特別是激酶和磷酸酶的參與。[72]酶也能產生運動,通過催化肌球蛋白上ATP的水解產生肌肉收縮,並且能夠作為細胞骨架的一部分參與運送胞內物質。[73]一些位於細胞膜上的ATP酶作為離子泵參與主動運輸。一些生物體中比較奇特的功能也有酶的參與,例如螢光素酶可以為螢火蟲發光。[74]病毒中也含有酶,或參與侵染細胞(如HIV整合酶和反轉錄酶),或參與病毒顆粒從宿主細胞的釋放(如流感病毒的神經胺酸酶)。
酶的一個非常重要的功能是參與在動物消化系統的工作。以澱粉酶和蛋白酶為代表的一些酶可以將進入消化道的大分子(澱粉和蛋白質)降解為小分子,以便於腸道吸收。澱粉不能被腸道直接吸收,而酶可以將澱粉水解為麥芽糖或更進一步水解為葡萄糖等腸道可以吸收的小分子。不同的酶分解不同的食物受質。在草食性反芻動物的消化系統中存在一些可以產生纖維素酶的細菌,纖維素酶可以分解植物細胞壁中的纖維素,從而提供可被吸收的養料。[75]
代謝[編輯]

多個酶以某一特定的順序發揮功能,共同構成了代謝途徑。在代謝途徑中,前一個酶的產物是後一個酶的受質;每個酶催化反應後,產物被傳遞到另一個酶。有些情況下,不同的酶可以平行地催化同一個反應,從而允許進行更為複雜的調控:比如一個酶可以以較低的活性持續地催化該反應,而另一個酶在被誘導後可以較高的活性進行催化。
酶的存在確定了整個代謝按正確的途徑進行;而一旦沒有酶的存在,代謝既不能按所需步驟進行,也無法以足夠的速度完成合成以滿足細胞的需要。實際上如果沒有酶,代謝途徑,如糖解,無法獨立進行。例如,葡萄糖可以直接與ATP反應使得其一個或多個碳原子被磷酸化;在沒有酶的催化時,這個反應進行得非常緩慢以致可以忽略;而一旦加入己醣激酶,在6位上的碳原子的磷酸化反應獲得極大加速,雖然其他碳原子的磷酸化反應也在緩慢進行,但在一段時間後檢測可以發現,絕大多數產物為葡萄糖-6-磷酸。[76]於是每個細胞就可以通過這樣一套功能性酶來完成代謝途徑的整個反應網絡。
活性控制[編輯]
細胞內有五種控制酶催化活性的機制:
- 根據外界環境的變化,細胞可以增強或減弱酶的生產(即酶相關基因的轉錄和轉譯)。這屬於一種基因調控,被稱為酶的誘導和抑制。例如,當環境中出現如青黴素這樣的抗生素時,部分細菌可以對抗生素產生抗性,其原因就在於細菌體內的β-半乳醣苷酶被誘導而大量生產,這種酶可以水解青黴素分子上關鍵的β-乳胺環。另一個例子是在人體肝臟中存在一類酶對於藥物代謝非常重要的酶,細胞色素P450;對這一類酶的誘導或抑制,會導致藥物相互作用。
- 通過將特定的酶分隔在特定的細胞組分中,細胞可以完成不同的代謝途徑。例如,脂肪酸的合成是由細胞溶質、內質網和高基氏體中的一系列酶所完成,而脂肪酸的降解(以提供能量)是在粒線體中由另一系列酶通過β-氧化來完成。[77]
- 酶可以被抑制劑與活化劑所調控。例如,一個代謝途徑中的終產物常常是這一途徑中第一個酶的抑制劑,從而調控這一代謝途徑的產物量。這種調控機制被稱為負反饋機制,因為終產物的合成量是受其自身濃度調控。負反饋機制可以根據細胞的需要,有效地調節中間代謝物的合成速率,從而使細胞的能量和物質的分配更為高效,並防止多餘產物的合成。控制酶的作用,可以在生物體內維持一個穩定的內部環境(即體內平衡)。
- 轉譯後修飾也可以調控酶的活性。這些修飾包括磷酸化、肉豆蔻酸化和醣基化。例如,細胞接受胰島素信號後,對包括肝糖合酶在內的多個酶進行磷酸化,幫助控制肝糖的合成或降解,使得細胞可以對血糖的變化產生反應。[78]另一個轉譯後修飾的例子是多肽鏈的剪切。胰凝乳蛋白酶,一種消化性蛋白酶,是產生於胰臟中的無活性的胰凝乳蛋白酶原,這一蛋白通過運輸到達胃後才被活化。這種方式有效地防止了胰凝乳蛋白酶在進入腸之前消化胰臟或其他組織。這種無活性的酶的前體被命名為酶原。
- 還有一些酶可以通過定位到不同環境後而被活化,比如從還原態的環境(細胞質)到氧化態環境(細胞周質空間),從高pH環境到低pH環境等。流感病毒的紅血球凝集素蛋白就是一個例子:當它接觸到宿主細胞囊泡的酸性環境時,它的構象立刻發生變化,導致其獲得活化。[79]
相關疾病[編輯]

酶的活性必須嚴格控制以維持體內平衡,對於能夠影響一個關鍵酶的功能的任何基因缺陷(如突變導致活性變化,過量表現、過低表現或刪除突變)都可能導致遺傳性疾病發生。許多事實顯示,一種致命疾病的病因可以只是由於人體中的數千種酶中的一種發生功能故障。
- 苯丙酮尿症:此種病症是典型的酶相關病例之一。病因是苯丙胺酸羥化酶(其功能是催化苯丙胺酸降解過程中的第一步)上一個胺基酸位點發生了突變,導致體內苯丙胺酸和相關產物的水平過高,如果沒有得到合適的治療,會進一步導致智能障礙。[80]
- 紫質病:該病是由於血基質生物合成途徑中特定酶的酶活性過低(基因突變或其他原因導致),使得中間產物紫質的產生和排泄異常,在一定誘因(如陽光照射)下,可導致皮膚或其他組織器官發生病變。[81]
- 當生殖細胞中編碼DNA修復相關酶的基因發生突變,其結果會導致遺傳性癌症綜合病徵,如著色性干皮症。[82]DNA修復酶的缺陷導致人體喪失修復突變基因的能力。發生的突變不斷積累,最終使得患者有多種癌症發生。
酶的口服給藥,可用於治療多種疾病(如胰腺功能不全和乳糖不耐受症)。由於酶作為蛋白質可能在消化道環境中失活或被降解,因此一種非侵入性的成像方法被開發用於監測作為藥物的酶在消化道中的活性變化。[83]
工業應用[編輯]
酶因爲能高效催化特定反應,已在化工等行業得到廣泛應用。總的來說,酶的應用因爲它們能催化的反應數目少、在有機溶劑中以及高溫環境下不穩定而受到限制。因此,酶工程這一熱門學科應運而生。酶工程旨在藉助合理的設計或體外進化的方法研發具有新特性的酶[84][85]。目前,酶工程學已取得了一定成果,研究人員甚至已「從頭」(即不以任何自然界中的酶爲模板)設計出了一些能催化在自然界中不能發生的反應的酶[86]。儘管酶催化的工業過程非常高效,但一些酶依賴於菸鹼醯胺輔因子(NADH/NAD+、NADP+/NAPH)。 由於此類輔助因子的高價格,這些工藝在經濟上不具有競爭力。 最近,一些合成化合物被認為是具有經濟前景的天然輔因子的仿生對應物[87]。
| 應用領域 | 酶 | 用途 |
|---|---|---|
| 生物燃料工業 | 纖維素酶 | 將纖維素分解可通過發酵轉化爲纖維素乙醇的糖[88] |
| 木質酶 | 對準備用於生物燃料生產的生物質進行預處理[88] | |
| 生物洗滌劑 | 蛋白酶、澱粉酶、脂肪酶 | 洗衣或清洗餐具時去除蛋白質、澱粉、脂肪或油漬[89] |
| 甘露聚醣酶 | 去除食物食品污漬[89] | |
| 釀酒業 | 澱粉酶、葡聚醣酶、蛋白酶 | 從麥芽中分解多醣及蛋白質[90]:150–9 |
| β-葡聚醣酶 | 提高麥汁和啤酒過濾特徵[90]:545 | |
| 澱粉葡萄糖苷酶及支鏈澱粉酶 | 製作低熱量啤酒及調整發酵特性[90]:575 | |
| 乙醯乳酸脫羧酶(ALDC) | 利用減少丁二酮形成來提昇發酵效率[91] | |
| 烹飪用 | 木瓜蛋白酶 | 使肉變嫰,容易烹飪[92] |
| 乳品業 | 凝乳酶 | 在奶酪生產過程中酸化蛋白質[93] |
| 脂酶 | 製作卡芒貝爾奶酪及像羅克福乾酪之類的藍乾酪[94] | |
| 食品加工 | 澱粉酶 | 從澱粉製造醣類,例如製作高果糖玉米糖漿[95] |
| 蛋白酶 | 降低麵粉中的蛋白質比例,例如用在餅乾製造中[96] | |
| 胰蛋白酶 | 製作防過敏的嬰兒食品[96] | |
| 纖維素酶、果膠酶 | 澄清果汁[97] | |
| 分子生物學 | 核酸酶、聚合酶、DNA連接酶 | 藉助限制性核酸內切酶以及PCR技術產生重組DNA[1]:6.2 |
| 造紙業 | 木聚醣酶、半纖維素酶及木質素過氧化物酶 | 從木漿中移除木質素[98] |
| 個人護理 | 蛋白酶 | 清除隱形眼鏡上的蛋白質,防止感染。[99] |
| 澱粉工業 | 澱粉酶 | 將澱粉轉換成葡萄糖及各種糖漿[100] |
醫藥應用[編輯]
由於六大類酵素只有水解酶在人體細胞外作用,其他均在細胞內發揮功能,所以大多數口服酵素都沒有證據有生理功效或病理療效。這是因為絕大部分的酵素都是蛋白質,而易受腸胃道酸鹼值及人體本身消化道的消化酵素破壞。目前有醫療上較可靠證據的,只有屬於水解酶的鳳梨酶,且必須藉由特殊劑型設計以腸溶衣保護,才能可能發揮療效。由於經口攝入的胜肽或蛋白質,只有二肽及三肽能完整地被腸胃道吸收進入血循,所以醫藥專家仍對此存疑。[101]
參見[編輯]
| 維基共享資源上的相關多媒體資源:酶 |
參考文獻[編輯]
- ^ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 Stryer L, Berg JM, Tymoczko JL. Biochemistry 5th. San Francisco: W.H. Freeman. 2002 [2016-08-16]. ISBN 0-7167-4955-6. (原始內容存檔於2016-01-05).

- ^ Schomburg I, Chang A, Placzek S, Söhngen C, Rother M, Lang M, Munaretto C, Ulas S, Stelzer M, Grote A, Scheer M, Schomburg D. BRENDA in 2013: integrated reactions, kinetic data, enzyme function data, improved disease classification: new options and contents in BRENDA. Nucleic Acids Research. January 2013, 41 (Database issue): D764–72. PMC 3531171
 . PMID 23203881. doi:10.1093/nar/gks1049.
. PMID 23203881. doi:10.1093/nar/gks1049.
- ^ Radzicka A, Wolfenden R. A proficient enzyme. Science. January 1995, 267 (5194): 90–931. Bibcode:1995Sci...267...90R. PMID 7809611. doi:10.1126/science.7809611.
- ^ Callahan BP, Miller BG. OMP decarboxylase—An enigma persists. Bioorganic Chemistry. December 2007, 35 (6): 465–9. PMID 17889251. doi:10.1016/j.bioorg.2007.07.004.
- ^ (法文)de Réaumur, RAF. Observations sur la digestion des oiseaux. Histoire de l'academie royale des sciences. 1752, 1752: 266, 461.
- ^ (英文)Williams, H. S.(1904)A History of Science: in Five Volumes. Volume IV: Modern Development of the Chemical and Biological Sciences (頁面存檔備份,存於網際網路檔案館) Harper and Brothers (New York) Accessed 04 April 2007
- ^ (英文)諾貝爾獎獲得者愛德華·比希納的簡歷 (頁面存檔備份,存於網際網路檔案館)Accessed 04 April 2007
- ^ (英文)愛德華·比希納在1907年的諾貝爾獎獲獎演說 (頁面存檔備份,存於網際網路檔案館)Accessed 04 April 2007
- ^ (英文)1946年度諾貝爾化學獎獲得者 (頁面存檔備份,存於網際網路檔案館)Accessed 04 April 2007
- ^ (英文)Blake CC, Koenig DF, Mair GA, North AC, Phillips DC, Sarma VR. Structure of hen egg-white lysozyme. A three-dimensional Fourier synthesis at 2 Angstrom resolution.. Nature. 1965, 22 (206): 757–761. PMID 5891407.
- ^ (英文)1989年度諾貝爾化學獎 (頁面存檔備份,存於網際網路檔案館)授予了托馬斯·切赫和雪梨·奧爾特曼以獎勵他們發現RNA分子的催化性質。
- ^ Nomenclature Committee. Classification and Nomenclature of Enzymes by the Reactions they Catalyse. International Union of Biochemistry and Molecular Biology (NC-IUBMB). School of Biological and Chemical Sciences, Queen Mary, University of London. [2016-08-16]. (原始內容存檔於2015-03-17).
- ^ Nomenclature Committee. EC 2.7.1.1. International Union of Biochemistry and Molecular Biology (NC-IUBMB). School of Biological and Chemical Sciences, Queen Mary, University of London. [2016-08-16]. (原始內容存檔於2014-12-01).
- ^ Anfinsen CB. Principles that govern the folding of protein chains. Science. July 1973, 181 (4096): 223–30. Bibcode:1973Sci...181..223A. PMID 4124164. doi:10.1126/science.181.4096.223.
- ^ Dunaway-Mariano D. Enzyme function discovery. Structure (London, England : 1993). November 2008, 16 (11): 1599–600. PMID 19000810. doi:10.1016/j.str.2008.10.001.
- ^ Petsko GA, Ringe D. Chapter 1: From sequence to structure. Protein structure and function. London: New Science. 2003: 27 [2016-08-16]. ISBN 978-1405119221. (原始內容存檔於2021-05-20).
- ^ Chen LH, Kenyon GL, Curtin F, Harayama S, Bembenek ME, Hajipour G, Whitman CP. 4-Oxalocrotonate tautomerase, an enzyme composed of 62 amino acid residues per monomer. The Journal of Biological Chemistry. September 1992, 267 (25): 17716–21. PMID 1339435.
- ^ Smith S. The animal fatty acid synthase: one gene, one polypeptide, seven enzymes. FASEB Journal. December 1994, 8 (15): 1248–59. PMID 8001737.
- ^ The Catalytic Site Atlas. The European Bioinformatics Institute. [4 April 2007]. (原始內容存檔於2016-06-04).
- ^ 20.0 20.1 Suzuki H. Chapter 7: Active Site Structure. How Enzymes Work: From Structure to Function. Boca Raton, FL: CRC Press. 2015: 117–140. ISBN 978-981-4463-92-8.
- ^ Krauss G. The Regulations of Enzyme Activity. Biochemistry of Signal Transduction and Regulation 3rd. Weinheim: Wiley-VCH. 2003: 89–114 [2016-08-16]. ISBN 9783527605767. (原始內容存檔於2021-05-20).
- ^ Jocelyn E.KREBS; et al. Gene XI. JONES&BARTLETT LEARNING(高等教育出版社出版). 2014. ISBN 978-7-04-039649-2.
- ^ Jaeger KE, Eggert T. Enantioselective biocatalysis optimized by directed evolution. Current Opinion in Biotechnology. August 2004, 15 (4): 305–13. PMID 15358000. doi:10.1016/j.copbio.2004.06.007.
- ^ Shevelev IV, Hübscher U. The 3' 5' exonucleases. Nature Reviews Molecular Cell Biology. May 2002, 3 (5): 364–76. PMID 11988770. doi:10.1038/nrm804.
- ^ Ibba M, Soll D. Aminoacyl-tRNA synthesis. Annual Review of Biochemistry. 2000, 69: 617–50. PMID 10966471. doi:10.1146/annurev.biochem.69.1.617.
- ^ Rodnina MV, Wintermeyer W. Fidelity of aminoacyl-tRNA selection on the ribosome: kinetic and structural mechanisms. Annual Review of Biochemistry. 2001, 70: 415–35. PMID 11395413. doi:10.1146/annurev.biochem.70.1.415.
- ^ Zenkin N, Yuzenkova Y, Severinov K. Transcript-assisted transcriptional proofreading. Science. July 2006, 313 (5786): 518–20. Bibcode:2006Sci...313..518Z. PMID 16873663. doi:10.1126/science.1127422.
- ^ Khersonsky O, Tawfik DS. Enzyme promiscuity: a mechanistic and evolutionary perspective. Annual Review of Biochemistry. 2010, 79: 471–505. PMID 20235827. doi:10.1146/annurev-biochem-030409-143718.
- ^ O'Brien PJ, Herschlag D. Catalytic promiscuity and the evolution of new enzymatic activities. Chemistry & Biology. April 1999, 6 (4): R91–R105. PMID 10099128. doi:10.1016/S1074-5521(99)80033-7.
- ^ Fischer E. Einfluss der Configuration auf die Wirkung der Enzyme [Influence of configuration on the action of enzymes]. Berichte der Deutschen chemischen Gesellschaft zu Berlin. 1894, 27 (3): 2985–93 [2007-09-27]. doi:10.1002/cber.18940270364. (原始內容存檔於2011-05-11) (德語). From page 2992: "Um ein Bild zu gebrauchen, will ich sagen, dass Enzym und Glucosid wie Schloss und Schlüssel zu einander passen müssen, um eine chemische Wirkung auf einander ausüben zu können." (To use an image, I will say that an enzyme and a glucoside [i.e., glucose derivative] must fit like a lock and key, in order to be able to exert a chemical effect on each other.)
- ^ Cooper GM. Chapter 2.2: The Central Role of Enzymes as Biological Catalysts. The Cell: a Molecular Approach 2nd. Washington (DC ): ASM Press. 2000 [2016-08-16]. ISBN 0-87893-106-6. (原始內容存檔於2021-05-20).
- ^ Koshland DE. Application of a Theory of Enzyme Specificity to Protein Synthesis. Proceedings of the National Academy of Sciences of the United States of America. February 1958, 44 (2): 98–104. Bibcode:1958PNAS...44...98K. PMC 335371
 . PMID 16590179. doi:10.1073/pnas.44.2.98.
. PMID 16590179. doi:10.1073/pnas.44.2.98.
- ^ Vasella A, Davies GJ, Böhm M. Glycosidase mechanisms. Current Opinion in Chemical Biology. October 2002, 6 (5): 619–29. PMID 12413546. doi:10.1016/S1367-5931(02)00380-0.
- ^ Boyer R. Chapter 6: Enzymes I, Reactions, Kinetics, and Inhibition. Concepts in Biochemistry 2nd. New York, Chichester, Weinheim, Brisbane, Singapore, Toronto.: John Wiley & Sons, Inc. 2002: 137–8. ISBN 0-470-00379-0. OCLC 51720783.
- ^ Savir Y, Tlusty T. Scalas E , 編. Conformational proofreading: the impact of conformational changes on the specificity of molecular recognition (PDF). PLoS ONE. 2007, 2 (5): e468 [2016-08-16]. Bibcode:2007PLoSO...2..468S. PMC 1868595
 . PMID 17520027. doi:10.1371/journal.pone.0000468. (原始內容 (PDF)存檔於2011-05-14).
. PMID 17520027. doi:10.1371/journal.pone.0000468. (原始內容 (PDF)存檔於2011-05-14).
- ^ Fersht A. Enzyme Structure and Mechanism. San Francisco: W.H. Freeman. 1985: 50–2. ISBN 0-7167-1615-1.
- ^ Warshel A, Sharma PK, Kato M, Xiang Y, Liu H, Olsson MH. Electrostatic basis for enzyme catalysis. Chemical Reviews. August 2006, 106 (8): 3210–35. PMID 16895325. doi:10.1021/cr0503106.
- ^ Cox MM, Nelson DL. Chapter 6.2: How enzymes work. Lehninger Principles of Biochemistry 6th. New York, N.Y.: W.H. Freeman. 2013: 195 [2016-08-16]. ISBN 978-1464109621. (原始內容存檔於2021-05-20).
- ^ Benkovic SJ, Hammes-Schiffer S. A perspective on enzyme catalysis. Science. August 2003, 301 (5637): 1196–202. Bibcode:2003Sci...301.1196B. PMID 12947189. doi:10.1126/science.1085515.
- ^ Jencks WP. Catalysis in Chemistry and Enzymology. Mineola, N.Y: Dover. 1987. ISBN 0-486-65460-5.
- ^ Villa J, Strajbl M, Glennon TM, Sham YY, Chu ZT, Warshel A. How important are entropic contributions to enzyme catalysis?. Proceedings of the National Academy of Sciences of the United States of America. October 2000, 97 (22): 11899–904. Bibcode:2000PNAS...9711899V. PMC 17266
 . PMID 11050223. doi:10.1073/pnas.97.22.11899.
. PMID 11050223. doi:10.1073/pnas.97.22.11899.
- ^ Ramanathan A, Savol A, Burger V, Chennubhotla CS, Agarwal PK. Protein conformational populations and functionally relevant substates. Acc. Chem. Res. 2014, 47 (1): 149–56. PMID 23988159. doi:10.1021/ar400084s.
- ^ Tsai CJ, Del Sol A, Nussinov R. Protein allostery, signal transmission and dynamics: a classification scheme of allosteric mechanisms. Mol Biosyst. 2009, 5 (3): 207–16. PMC 2898650
 . PMID 19225609. doi:10.1039/b819720b.
. PMID 19225609. doi:10.1039/b819720b.
- ^ Changeux JP, Edelstein SJ. Allosteric mechanisms of signal transduction. Science. June 2005, 308 (5727): 1424–8. Bibcode:2005Sci...308.1424C. PMID 15933191. doi:10.1126/science.1108595.
- ^ de Bolster MW. Glossary of Terms Used in Bioinorganic Chemistry: Cofactor. International Union of Pure and Applied Chemistry. 1997 [30 October 2007]. (原始內容存檔於2017-01-21).
- ^ Chapman-Smith A, Cronan JE. The enzymatic biotinylation of proteins: a post-translational modification of exceptional specificity. Trends Biochem. Sci. 1999, 24 (9): 359–63. PMID 10470036. doi:10.1016/s0968-0004(99)01438-3.
- ^ Fisher Z, Hernandez Prada JA, Tu C, Duda D, Yoshioka C, An H, Govindasamy L, Silverman DN, McKenna R. Structural and kinetic characterization of active-site histidine as a proton shuttle in catalysis by human carbonic anhydrase II. Biochemistry. February 2005, 44 (4): 1097–115. PMID 15667203. doi:10.1021/bi0480279.
- ^ 48.0 48.1 Wagner AL. Vitamins and Coenzymes. Krieger Pub Co. 1975. ISBN 0-88275-258-8.
- ^ BRENDA The Comprehensive Enzyme Information System. Technische Universität Braunschweig. [23 February 2015]. (原始內容存檔於2015-05-06).
- ^ Törnroth-Horsefield S, Neutze R. Opening and closing the metabolite gate. Proceedings of the National Academy of Sciences of the United States of America. December 2008, 105 (50): 19565–6. Bibcode:2008PNAS..10519565T. PMC 2604989
 . PMID 19073922. doi:10.1073/pnas.0810654106.
. PMID 19073922. doi:10.1073/pnas.0810654106.
- ^ (英文)Ferguson, S. J.; Nicholls, David; Ferguson, Stuart. Bioenergetics 3 3rd. San Diego: Academic. 2002. ISBN 0-12-518121-3.
- ^ (英文)Maren TH. Carbonic anhydrase: chemistry, physiology, and inhibition. Physiol Rev. 1967, 47 (4): 595–781. PMID 4964060.
- ^ (法文)Henri,V. Theorie generale de l'action de quelques diastases. Compt. rend. hebd. Acad. Sci. Paris. 1902, 135: 916–919.
- ^ (德文)Michaelis L., Menten M. Die Kinetik der Invertinwirkung. Biochem. Z. 1913, 49: 333–369. English translation (頁面存檔備份,存於網際網路檔案館) Accessed 6 April 2007
- ^ (英文)Radzicka A, Wolfenden R. A proficient enzyme.. Science. 1995, 6 (267): 90–931. PMID 7809611.
- ^ (英文)Ellis RJ. Macromolecular crowding: obvious but underappreciated. Trends Biochem. Sci. 2001, 26 (10): 597–604. PMID 11590012.
- ^ (英文)Kopelman R. Fractal Reaction Kinetics. Science. 1988, 241 (4873): 1620–26. doi:10.1126/science.241.4873.1620.
- ^ (英文)Savageau MA. Michaelis-Menten mechanism reconsidered: implications of fractal kinetics. J. Theor. Biol. 1995, 176 (1): 115–24. PMID 7475096.
- ^ (英文)Schnell S, Turner TE. Reaction kinetics in intracellular environments with macromolecular crowding: simulations and rate laws. Prog. Biophys. Mol. Biol. 2004, 85 (2–3): 235–60. PMID 15142746.
- ^ (英文)Xu F, Ding H. A new kinetic model for heterogeneous (or spatially confined) enzymatic catalysis: Contributions from the fractal and jamming (overcrowding) effects. Appl. Catal. A: Gen. 2007, 317 (1): 70–81. doi:10.1016/j.apcata.2006.10.014.
- ^ (英文)Garcia-Viloca M., Gao J., Karplus M., Truhlar D. G. How enzymes work: analysis by modern rate theory and computer simulations.. Science. 2004, 303 (5655): 186–195. PMID 14716003.
- ^ (英文)Olsson M. H., Siegbahn P. E., Warshel A. Simulations of the large kinetic isotope effect and the temperature dependence of the hydrogen atom transfer in lipoxygenase. J. Am. Chem. Soc. 2004, 126 (9): 2820–1828. PMID 14995199.
- ^ (英文)Masgrau L., Roujeinikova A., Johannissen L. O., Hothi P., Basran J., Ranaghan K. E., Mulholland A. J., Sutcliffe M. J., Scrutton N. S., Leys D. Atomic Description of an Enzyme Reaction Dominated by Proton Tunneling. Science. 2006, 312 (5771): 237–241. PMID 16614214.
- ^ (英文)Kohen, A., Cannio, R., Bartolucci, S., Klinman, J. P. Enzyme dynamics and hydrogen tunnelling in a thermophilic alcohol dehydrogenase. Nature. 1999, 399 (6735): 496–9. PMID 10365965.
- ^ (英文)Ball, P. Enzymes: by chance, or by design?. Nature. 2004, 431 (7007): 396–7. PMID 15385982.
- ^ Laurence A. Moran, Robert Horton, Gray Scrimgeour & Marc D. Perry. Properties of Enzymes. Principles of Biochemistry (Fifth Edition) [生物化學原理]. 培生出版集團. 2011: 148–152. ISBN 0-321-70733-8.
- ^ (英文)Cleland, W.W. The Kinetics of Enzyme-catalyzed Reactions with two or more Substrates or Products 2. {I}nhibition: Nomenclature and Theory. Biochim. Biophys. Acta. 1963, 67: 173–187.
- ^ (英文)Price, NC. What is meant by 'competitive inhibition'?. Trends in Biochemical Sciences. 1979, 4 (11): pN272. doi:10.1016/0968-0004(79)90205-6.
- ^ (英文)Poulin R, Lu L, Ackermann B, Bey P, Pegg AE. Mechanism of the irreversible inactivation of mouse ornithine decarboxylase by alpha-difluoromethylornithine. Characterization of sequences at the inhibitor and coenzyme binding sites. (頁面存檔備份,存於網際網路檔案館) J Biol Chem. 1992 Jan 5;267 (1):150–8. PMID 1730582
- ^ (英文)Ball, Philip (2006) The Devil's Doctor: Paracelsus and the World of Renaissance Magic and Science. Farrar, Straus and Giroux ISBN 978-0-374-22979-5
- ^ (英文)Yoshikawa S and Caughey WS. Infrared evidence of cyanide binding to iron and copper sites in bovine heart cytochrome c oxidase. Implications regarding oxygen reduction.. J Biol Chem. May 1990, 265 (14): 7945–7958 [2007-10-17]. PMID 2159465. (原始內容存檔於2008-09-25).
- ^ (英文)Hunter T. Protein kinases and phosphatases: the yin and yang of protein phosphorylation and signaling.. Cell. 1995, 80 (2): 225–236. PMID 7834742.
- ^ Berg JS, Powell BC, Cheney RE. A millennial myosin census.. Mol Biol Cell. 2001, 12 (4): 780–794. PMID 11294886.
- ^ (英文)Meighen EA. Molecular biology of bacterial bioluminescence.. Microbiol Rev. 1991, 55 (1): 123–142. PMID 2030669.
- ^ (英文)Mackie RI, White BA; White. Recent advances in rumen microbial ecology and metabolism: potential impact on nutrient output. J. Dairy Sci. 1 October 1990, 73 (10): 2971–95. PMID 2178174. doi:10.3168/jds.S0022-0302(90)78986-2.
- ^ (英文)Jennifer McDowall. Enzymes of Glycolysis.. [2015-01-23]. (原始內容存檔於2021-05-20).
- ^ (英文)Faergeman N. J, Knudsen J. Role of long-chain fatty acyl-CoA esters in the regulation of metabolism and in cell signalling. Biochem J. April 1997, 323: 1–12. PMID 9173866.
- ^ (英文)Doble B. W., Woodgett J. R. GSK-3: tricks of the trade for a multi-tasking kinase. J. Cell. Sci. April 2003, 116: 1175–1186 [2007-10-07]. PMID 12615961. (原始內容存檔於2007-09-30).
- ^ (英文)Ciampor F, Cmarko D, Cmarkova J, Zavodska E. Influenza virus M2 protein and haemagglutinin conformation changes during intracellular transport.. Acta Virol. 1995, 39 (3): 171 – 181. PMID 8579000.
- ^ 預防醫學基金會. 苯酮尿症. [2015-01-23]. (原始內容存檔於2015-02-16).
- ^ 台大醫院. 紫質症. [2015-01-23]. (原始內容存檔於2015-02-16).
- ^ 謝紹安, 祝芷. 着色性干皮症. 眼科新進展. 1984, 3 [2016-09-28]. (原始內容存檔於2021-05-20).
- ^ (英文)Fuhrmann G, Leroux JC; Leroux. In vivo fluorescence imaging of exogenous enzyme activity in the gastrointestinal tract. Proceedings of the National Academy of Sciences. 2011, 108 (22): 9032–9037. PMC 3107327
 . PMID 21576491. doi:10.1073/pnas.1100285108.
. PMID 21576491. doi:10.1073/pnas.1100285108.
- ^ Renugopalakrishnan V, Garduño-Juárez R, Narasimhan G, Verma CS, Wei X, Li P. Rational design of thermally stable proteins: relevance to bionanotechnology. Journal of Nanoscience and Nanotechnology. November 2005, 5 (11): 1759–1767. PMID 16433409. doi:10.1166/jnn.2005.441.
- ^ Hult K, Berglund P. Engineered enzymes for improved organic synthesis. Current Opinion in Biotechnology. August 2003, 14 (4): 395–400. PMID 12943848. doi:10.1016/S0958-1669(03)00095-8.
- ^ Jiang L, Althoff EA, Clemente FR, Doyle L, Röthlisberger D, Zanghellini A, Gallaher JL, Betker JL, Tanaka F, Barbas CF, Hilvert D, Houk KN, Stoddard BL, Baker D. De novo computational design of retro-aldol enzymes. Science. March 2008, 319 (5868): 1387–91. Bibcode:2008Sci...319.1387J. PMC 3431203
 . PMID 18323453. doi:10.1126/science.1152692.
. PMID 18323453. doi:10.1126/science.1152692.
- ^ Characterization of Biomimetic Cofactors According to Stability, Redox Potentials, and Enzymatic Conversion by NADH Oxidase from Lactobacillus pentosus, ChemBioChem, 2017, 18(19):1944-1949 https://doi.org/10.1002/cbic.201700258
- ^ 88.0 88.1 Sun Y, Cheng J. Hydrolysis of lignocellulosic materials for ethanol production: a review. Bioresource Technology. May 2002, 83 (1): 1–11. PMID 12058826. doi:10.1016/S0960-8524(01)00212-7.
- ^ 89.0 89.1 Kirk O, Borchert TV, Fuglsang CC. Industrial enzyme applications. Current Opinion in Biotechnology. August 2002, 13 (4): 345–351. PMID 12323357. doi:10.1016/S0958-1669(02)00328-2.
- ^ 90.0 90.1 90.2 Briggs DE. Malts and Malting 1st. London: Blackie Academic. 1998. ISBN 978-0412298004.
- ^ Dulieu C, Moll M, Boudrant J, Poncelet D. Improved performances and control of beer fermentation using encapsulated alpha-acetolactate decarboxylase and modeling. Biotechnology Progress. 2000, 16 (6): 958–65. PMID 11101321. doi:10.1021/bp000128k.
- ^ Tarté R. Ingredients in Meat Products Properties, Functionality and Applications. New York: Springer. 2008: 177. ISBN 978-0-387-71327-4.
- ^ Chymosin – GMO Database. GMO Compass. European Union. 10 July 2010 [1 March 2015]. (原始內容存檔於2015-03-26).
- ^ Molimard P, Spinnler HE. Review: Compounds Involved in the Flavor of Surface Mold-Ripened Cheeses: Origins and Properties. Journal of Dairy Science. February 1996, 79 (2): 169–184. doi:10.3168/jds.S0022-0302(96)76348-8.
- ^ Guzmán-Maldonado H, Paredes-López O. Amylolytic enzymes and products derived from starch: a review. Critical Reviews in Food Science and Nutrition. September 1995, 35 (5): 373–403. PMID 8573280. doi:10.1080/10408399509527706.
- ^ 96.0 96.1 Protease – GMO Database. GMO Compass. European Union. 10 July 2010 [28 February 2015]. (原始內容存檔於2015-02-24).
- ^ Alkorta I, Garbisu C, Llama MJ, Serra JL. Industrial applications of pectic enzymes: a review. Process Biochemistry. January 1998, 33 (1): 21–28. doi:10.1016/S0032-9592(97)00046-0.
- ^ Bajpai P. Application of enzymes in the pulp and paper industry. Biotechnology Progress. March 1999, 15 (2): 147–157. PMID 10194388. doi:10.1021/bp990013k.
- ^ Begley CG, Paragina S, Sporn A. An analysis of contact lens enzyme cleaners. Journal of the American Optometric Association. March 1990, 61 (3): 190–4. PMID 2186082.
- ^ Farris PL. Economic Growth and Organization of the U.S. Starch Industry. BeMiller JN, Whistler RL (編). Starch Chemistry and Technology 3rd. London: Academic. 2009. ISBN 9780080926551.
- ^ 存档副本. [2021-03-23]. (原始內容存檔於2021-05-20).
100.https://dict.revised.moe.edu.tw/dictView.jsp?ID=1123&q=1&word=酶《教育部重編修訂本第六版》音:ㄇㄟˊ
拓展閱讀[編輯]
|
總論
詞源與歷史
|
酶結構及作用機理
動力學及抑制
|
| |||||||||||||||||||||||||||
| ||||||||||||